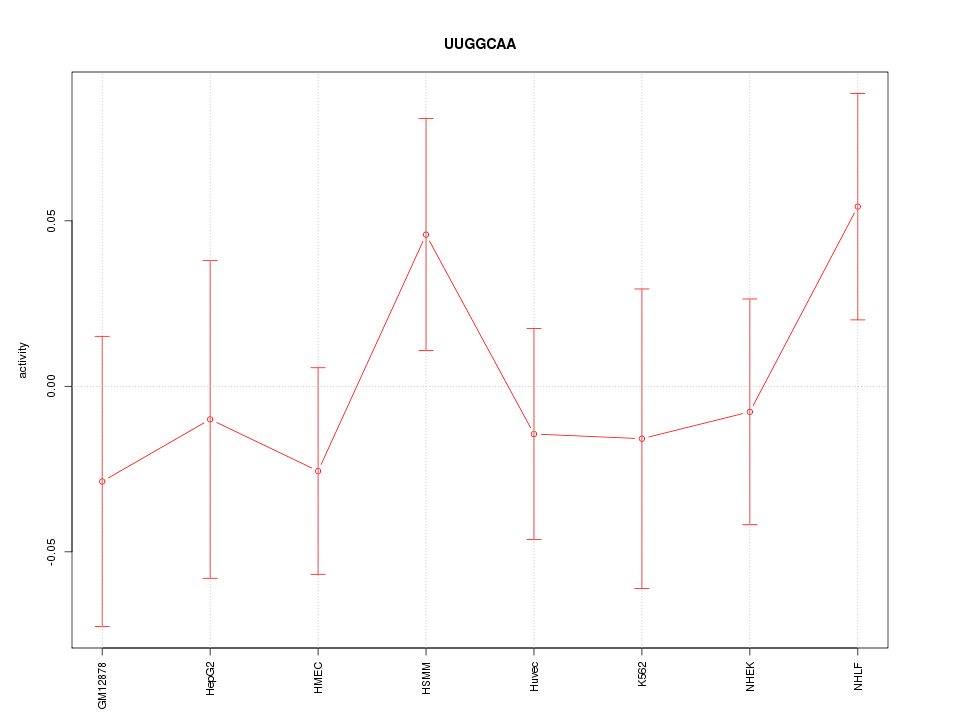
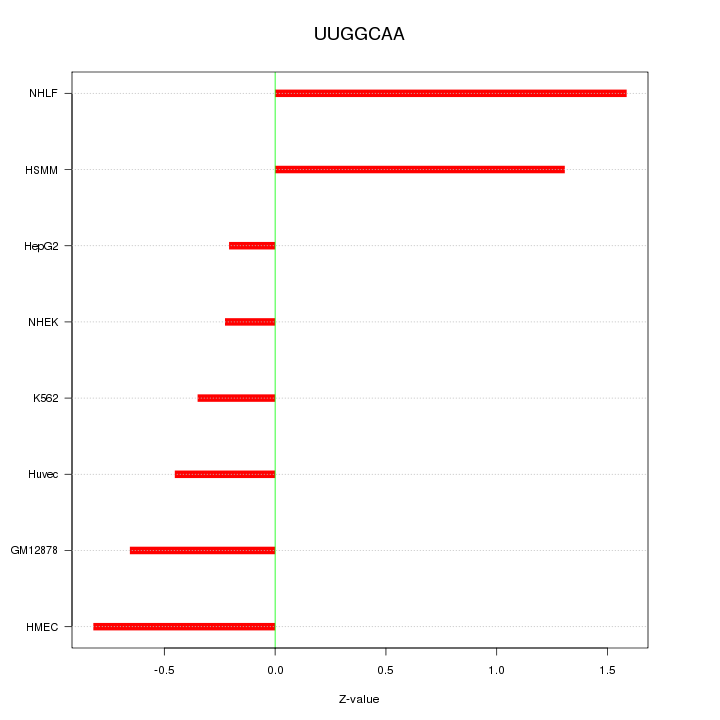
| 0.2 |
1.2 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.2 |
0.8 |
GO:0045112 |
integrin biosynthetic process(GO:0045112) |
| 0.2 |
1.3 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.4 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.1 |
0.8 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
1.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
1.3 |
GO:0000052 |
citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.1 |
0.2 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.1 |
0.3 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 |
0.2 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 |
0.2 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 |
0.4 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.1 |
GO:0021873 |
forebrain neuroblast division(GO:0021873) neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.1 |
0.3 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.5 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 |
0.2 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 |
0.3 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 |
0.5 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.1 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.1 |
0.4 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 |
0.9 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.1 |
0.2 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.1 |
0.7 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.0 |
0.3 |
GO:0010561 |
negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 |
0.2 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 |
0.4 |
GO:0042249 |
establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 |
0.1 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.4 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.0 |
0.2 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.8 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
1.4 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
0.5 |
GO:0021513 |
spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 |
0.4 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.5 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.2 |
GO:0097576 |
vacuole fusion(GO:0097576) |
| 0.0 |
0.1 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.3 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.1 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.2 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.2 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.5 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.0 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
0.1 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 |
0.1 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.0 |
0.6 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.3 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.2 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.0 |
0.1 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.1 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.2 |
GO:0015780 |
nucleotide-sugar transport(GO:0015780) |
| 0.0 |
0.2 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.0 |
0.2 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.1 |
GO:0007109 |
obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.0 |
0.1 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.0 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.0 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.2 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
0.1 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
0.3 |
GO:0002040 |
sprouting angiogenesis(GO:0002040) |
| 0.0 |
0.0 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.0 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.0 |
0.4 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.9 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
0.0 |
GO:0043558 |
regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.4 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 |
0.1 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.0 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.0 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 |
0.3 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.1 |
GO:1903729 |
regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of plasma membrane organization(GO:1903729) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 |
0.2 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |