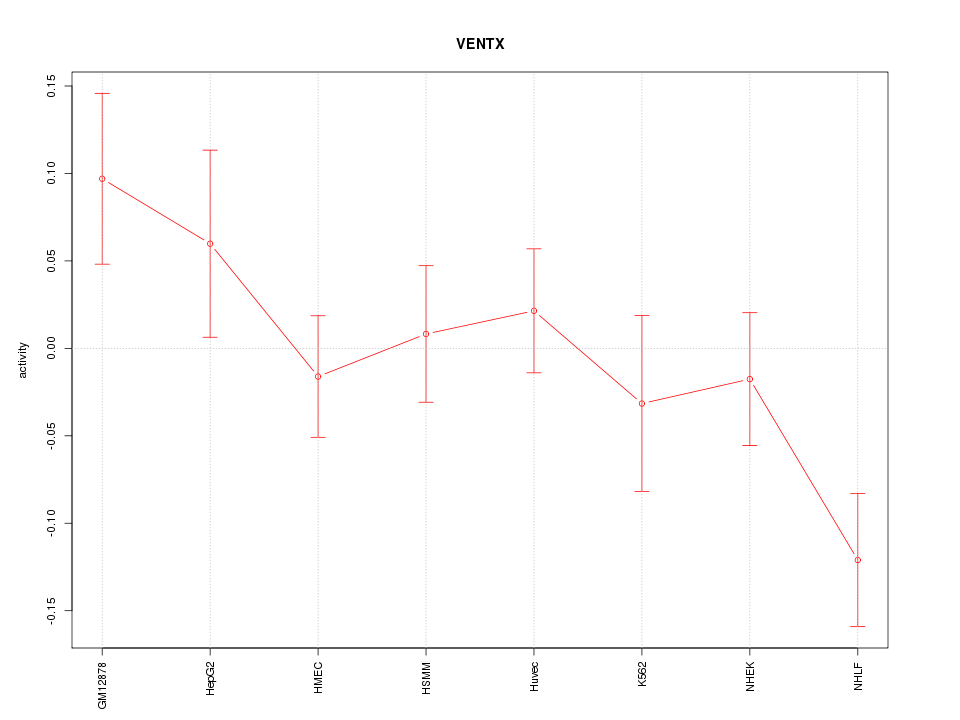
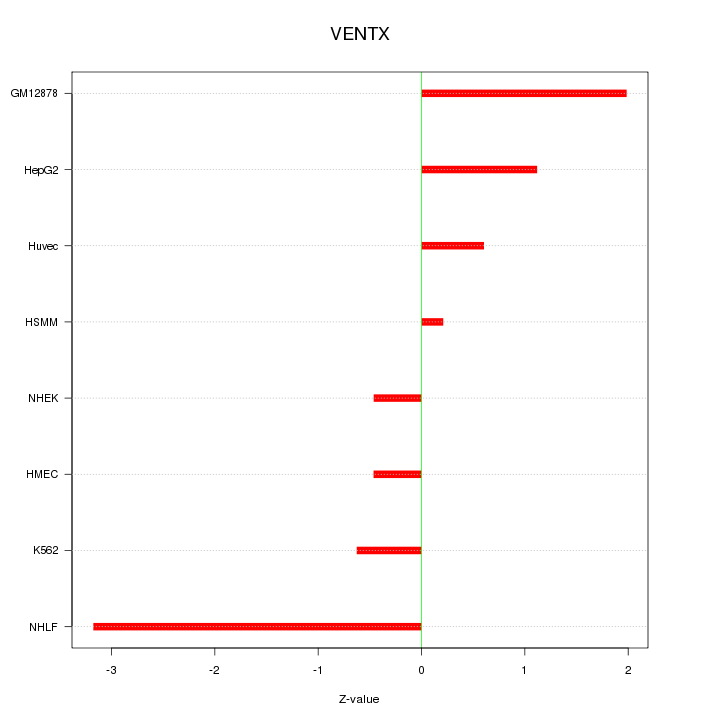
Motif ID: VENTX
Z-value: 1.437
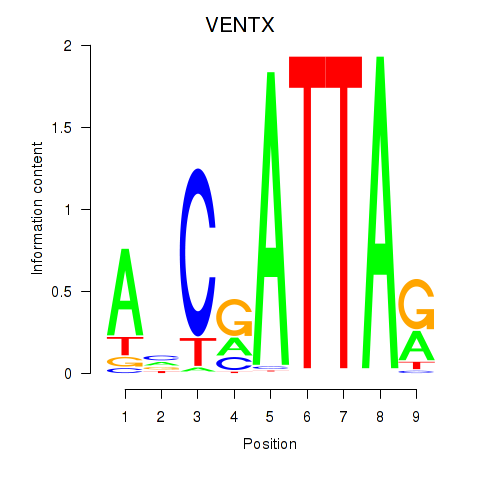
Transcription factors associated with VENTX:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| VENTX | ENSG00000151650.7 | VENTX |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 3.8 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.2 | 1.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 0.9 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.2 | 1.2 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.1 | 1.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.4 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.5 | GO:0060677 | ureteric bud elongation(GO:0060677) renal vesicle formation(GO:0072033) |
| 0.1 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 1.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.4 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.1 | 0.2 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.1 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.4 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 2.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.5 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0060235 | forebrain anterior/posterior pattern specification(GO:0021797) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 1.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.1 | 1.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.9 | GO:0008907 | integrase activity(GO:0008907) |
| 0.2 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.4 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.7 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.2 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 2.1 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) SAM domain binding(GO:0032093) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.1 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 1.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |