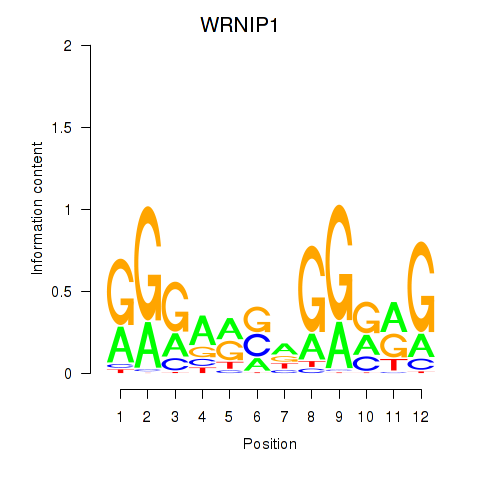
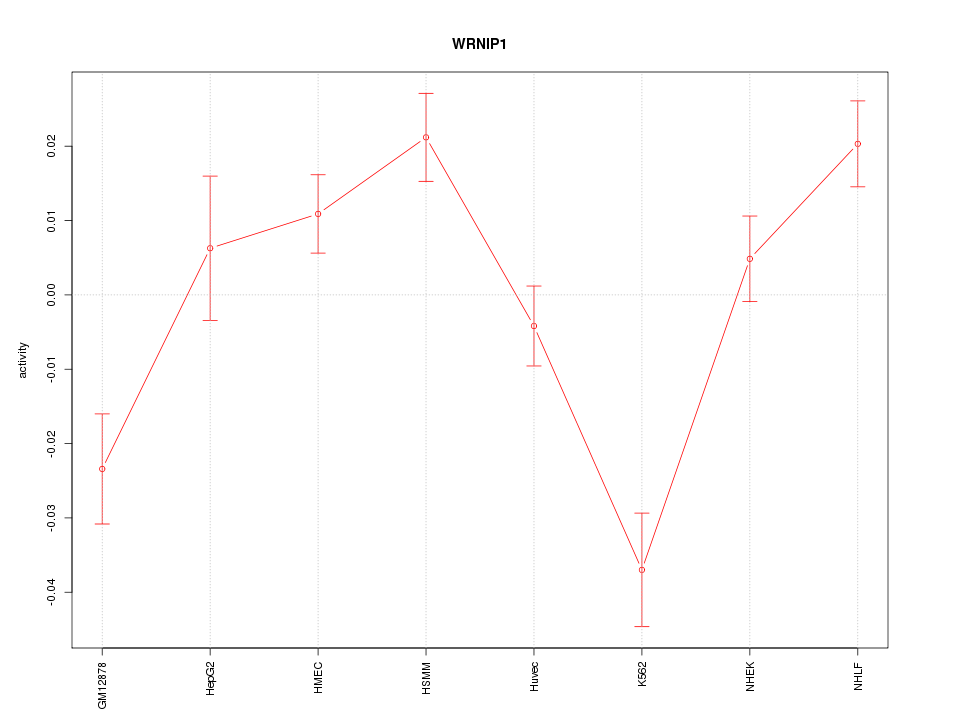
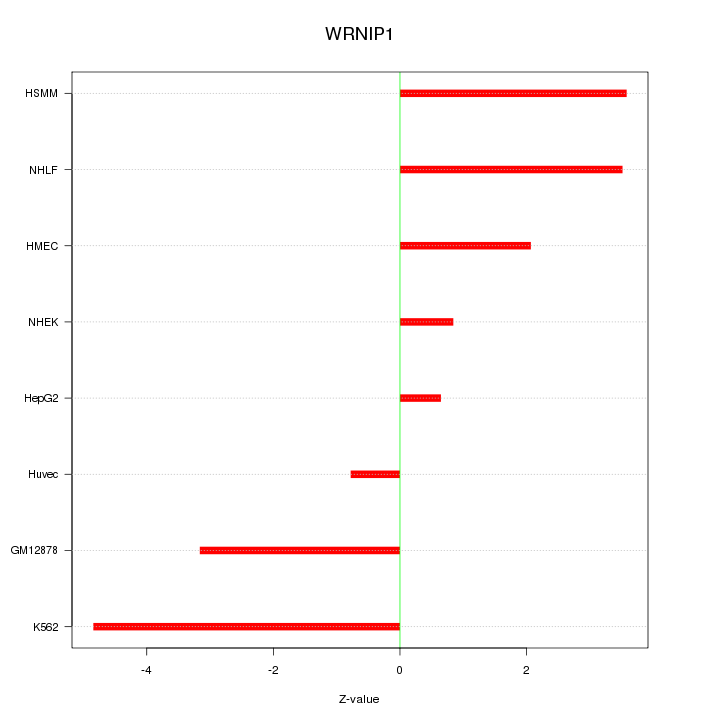
| 2.6 |
2.6 |
GO:0055098 |
response to low-density lipoprotein particle(GO:0055098) |
| 2.4 |
9.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 2.3 |
6.9 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 2.2 |
2.2 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 2.0 |
5.9 |
GO:2000064 |
regulation of cortisol biosynthetic process(GO:2000064) |
| 1.8 |
12.6 |
GO:2000111 |
positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.7 |
5.1 |
GO:0060599 |
lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 1.7 |
8.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 1.6 |
4.8 |
GO:0003308 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 1.5 |
4.6 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.5 |
6.1 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 1.3 |
4.0 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.3 |
3.9 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.2 |
7.4 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 1.2 |
3.6 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 1.2 |
1.2 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 1.2 |
5.9 |
GO:0007614 |
short-term memory(GO:0007614) |
| 1.2 |
3.5 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 1.2 |
3.5 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.1 |
16.1 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 1.0 |
6.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 1.0 |
3.0 |
GO:0046882 |
negative regulation of gonadotropin secretion(GO:0032277) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 1.0 |
4.0 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 1.0 |
9.9 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.9 |
7.6 |
GO:0070141 |
response to UV-A(GO:0070141) |
| 0.9 |
4.6 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.9 |
2.7 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.9 |
4.5 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.9 |
6.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.9 |
3.5 |
GO:0033986 |
response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 0.9 |
1.8 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.9 |
9.7 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.9 |
5.2 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.9 |
6.0 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.9 |
6.9 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.8 |
3.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.8 |
0.8 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.8 |
2.5 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.8 |
7.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.8 |
2.4 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.8 |
4.0 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.8 |
1.6 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.8 |
3.1 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.8 |
3.9 |
GO:0070884 |
calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.8 |
11.6 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.8 |
2.3 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.8 |
8.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 |
2.3 |
GO:0010881 |
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.8 |
2.3 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.8 |
0.8 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.8 |
5.3 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.7 |
2.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.7 |
3.7 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.7 |
2.2 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.7 |
5.2 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.7 |
2.9 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.7 |
3.6 |
GO:0044854 |
membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.7 |
2.2 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.7 |
2.2 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.7 |
2.2 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.7 |
5.0 |
GO:0015904 |
tetracycline transport(GO:0015904) |
| 0.7 |
4.3 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.7 |
3.5 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.7 |
2.1 |
GO:0071476 |
response to vitamin B1(GO:0010266) cellular hypotonic response(GO:0071476) |
| 0.7 |
3.4 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.7 |
2.1 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.7 |
6.6 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.7 |
7.2 |
GO:0060390 |
regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.6 |
2.5 |
GO:0001969 |
activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.6 |
1.9 |
GO:0060219 |
camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.6 |
0.6 |
GO:0030852 |
regulation of granulocyte differentiation(GO:0030852) |
| 0.6 |
1.9 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.6 |
2.5 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.6 |
3.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.6 |
3.6 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.6 |
1.2 |
GO:0010159 |
specification of organ position(GO:0010159) |
| 0.6 |
2.4 |
GO:0072048 |
pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.6 |
2.9 |
GO:0060346 |
bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.6 |
2.9 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 |
1.7 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.6 |
1.7 |
GO:1903115 |
regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.6 |
0.6 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.6 |
3.9 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.6 |
1.7 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.5 |
2.2 |
GO:0060209 |
estrus(GO:0060209) |
| 0.5 |
3.8 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.5 |
3.2 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 |
3.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.5 |
2.1 |
GO:0014742 |
positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.5 |
0.5 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.5 |
2.1 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.5 |
2.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 |
1.6 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.5 |
4.2 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.5 |
2.6 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.5 |
2.1 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.5 |
1.0 |
GO:0010903 |
negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.5 |
14.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.5 |
2.5 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.5 |
0.5 |
GO:0030202 |
heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.5 |
4.5 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.5 |
2.5 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) |
| 0.5 |
0.5 |
GO:0031223 |
auditory behavior(GO:0031223) |
| 0.5 |
7.3 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.5 |
1.5 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.5 |
5.3 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.5 |
2.9 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.5 |
1.4 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.5 |
1.4 |
GO:0071529 |
cementum mineralization(GO:0071529) |
| 0.5 |
2.8 |
GO:0097435 |
extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.5 |
1.4 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.5 |
1.8 |
GO:0015891 |
iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.5 |
1.4 |
GO:0032417 |
positive regulation of sodium:proton antiporter activity(GO:0032417) positive regulation of sodium ion transmembrane transport(GO:1902307) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.5 |
0.9 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.5 |
4.1 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.5 |
2.3 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.5 |
0.9 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 |
1.8 |
GO:0060037 |
pharyngeal system development(GO:0060037) |
| 0.4 |
0.9 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.4 |
1.3 |
GO:0010901 |
regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.4 |
2.2 |
GO:0006927 |
obsolete transformed cell apoptotic process(GO:0006927) |
| 0.4 |
1.8 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.4 |
2.6 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.4 |
5.2 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.4 |
2.1 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.4 |
1.3 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.4 |
3.0 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.4 |
1.7 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.4 |
1.7 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.4 |
2.1 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.4 |
8.2 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.4 |
1.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.4 |
2.4 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.4 |
2.0 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.4 |
3.2 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.4 |
1.2 |
GO:0071071 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.4 |
1.2 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.4 |
1.2 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.4 |
0.4 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.4 |
2.0 |
GO:1902743 |
regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.4 |
1.2 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.4 |
1.6 |
GO:0046108 |
uridine metabolic process(GO:0046108) |
| 0.4 |
0.8 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.4 |
2.0 |
GO:0010561 |
negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.4 |
1.6 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) regulation of cardiocyte differentiation(GO:1905207) |
| 0.4 |
2.4 |
GO:0030449 |
negative regulation of humoral immune response(GO:0002921) regulation of complement activation(GO:0030449) negative regulation of complement activation(GO:0045916) regulation of protein activation cascade(GO:2000257) negative regulation of protein activation cascade(GO:2000258) |
| 0.4 |
0.4 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.4 |
0.4 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.4 |
2.3 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 |
3.0 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.4 |
1.5 |
GO:0009296 |
obsolete flagellum assembly(GO:0009296) |
| 0.4 |
1.5 |
GO:1903170 |
negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of calcium ion transmembrane transport(GO:1903170) negative regulation of cation channel activity(GO:2001258) |
| 0.4 |
1.1 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.4 |
1.5 |
GO:0060343 |
trabecula formation(GO:0060343) trabecula morphogenesis(GO:0061383) |
| 0.4 |
2.6 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.4 |
1.5 |
GO:0042335 |
cuticle development(GO:0042335) |
| 0.4 |
1.5 |
GO:0072180 |
nephric duct morphogenesis(GO:0072178) mesonephric duct morphogenesis(GO:0072180) |
| 0.4 |
0.7 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.4 |
1.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.4 |
0.4 |
GO:0055009 |
atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.4 |
1.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) extracellular matrix assembly(GO:0085029) |
| 0.4 |
3.5 |
GO:0042749 |
circadian sleep/wake cycle process(GO:0022410) regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.3 |
2.1 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.3 |
1.0 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 |
1.0 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.3 |
3.1 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.3 |
0.7 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.3 |
1.0 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.3 |
0.7 |
GO:0048318 |
axial mesoderm development(GO:0048318) axial mesoderm morphogenesis(GO:0048319) |
| 0.3 |
0.7 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.3 |
1.3 |
GO:0060839 |
radial pattern formation(GO:0009956) lymphatic endothelial cell fate commitment(GO:0060838) endothelial cell fate commitment(GO:0060839) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 |
1.0 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.3 |
2.7 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 |
1.3 |
GO:0003197 |
endocardial cushion development(GO:0003197) |
| 0.3 |
1.0 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.3 |
1.0 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.3 |
1.0 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.3 |
0.6 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 |
1.0 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.3 |
7.9 |
GO:0043462 |
regulation of ATPase activity(GO:0043462) |
| 0.3 |
0.3 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.3 |
0.3 |
GO:0010193 |
response to ozone(GO:0010193) |
| 0.3 |
0.6 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.3 |
0.9 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.3 |
4.3 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.3 |
0.3 |
GO:0002686 |
negative regulation of leukocyte migration(GO:0002686) |
| 0.3 |
0.9 |
GO:1903960 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.3 |
6.3 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.3 |
2.1 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.3 |
0.3 |
GO:0022604 |
regulation of cell morphogenesis(GO:0022604) |
| 0.3 |
2.4 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.3 |
0.3 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 |
0.3 |
GO:0010044 |
response to aluminum ion(GO:0010044) |
| 0.3 |
0.9 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) negative regulation of receptor activity(GO:2000272) |
| 0.3 |
0.9 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.3 |
0.9 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.3 |
0.3 |
GO:0033590 |
response to cobalamin(GO:0033590) |
| 0.3 |
2.0 |
GO:0050818 |
regulation of coagulation(GO:0050818) |
| 0.3 |
0.3 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.3 |
1.1 |
GO:0010829 |
negative regulation of glucose transport(GO:0010829) |
| 0.3 |
0.6 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.3 |
1.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 |
1.7 |
GO:0032400 |
melanosome localization(GO:0032400) |
| 0.3 |
1.7 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 |
0.8 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.3 |
1.4 |
GO:0043587 |
tongue morphogenesis(GO:0043587) |
| 0.3 |
16.8 |
GO:0032387 |
negative regulation of intracellular transport(GO:0032387) |
| 0.3 |
2.8 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 |
1.4 |
GO:0002829 |
negative regulation of type 2 immune response(GO:0002829) |
| 0.3 |
4.4 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.3 |
0.3 |
GO:0030856 |
regulation of epithelial cell differentiation(GO:0030856) |
| 0.3 |
0.3 |
GO:0034372 |
triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 |
2.2 |
GO:0032429 |
regulation of phospholipase A2 activity(GO:0032429) |
| 0.3 |
1.6 |
GO:0048333 |
mesodermal cell fate commitment(GO:0001710) mesodermal cell differentiation(GO:0048333) |
| 0.3 |
1.6 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.3 |
3.0 |
GO:0043536 |
positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.3 |
3.2 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.3 |
18.2 |
GO:0030198 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.3 |
0.8 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.3 |
1.6 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 |
0.8 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.3 |
3.7 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 |
0.3 |
GO:2000178 |
negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 |
0.8 |
GO:0006210 |
pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) |
| 0.3 |
1.6 |
GO:0010812 |
negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.3 |
2.9 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.3 |
0.8 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.3 |
0.3 |
GO:0042640 |
anagen(GO:0042640) |
| 0.3 |
1.0 |
GO:0002331 |
immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.3 |
3.9 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 |
1.3 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.3 |
0.5 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.3 |
0.5 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.3 |
0.3 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 |
0.8 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 |
2.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.3 |
2.0 |
GO:0034394 |
protein localization to cell surface(GO:0034394) |
| 0.3 |
0.5 |
GO:0060746 |
maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.3 |
2.8 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.2 |
0.2 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 |
1.0 |
GO:0002335 |
mature B cell differentiation(GO:0002335) |
| 0.2 |
1.0 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.2 |
4.7 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.2 |
0.7 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.2 |
1.7 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.2 |
0.5 |
GO:0050665 |
hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.2 |
1.9 |
GO:0051957 |
positive regulation of amino acid transport(GO:0051957) |
| 0.2 |
6.0 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 |
0.7 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.2 |
0.5 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 |
0.5 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.2 |
13.0 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.2 |
0.9 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 |
1.8 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.2 |
0.7 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 |
0.9 |
GO:0030518 |
intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.2 |
4.6 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.2 |
0.7 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.2 |
0.7 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 |
0.2 |
GO:0046968 |
peptide antigen transport(GO:0046968) |
| 0.2 |
2.3 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.2 |
2.0 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 |
0.7 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 |
2.0 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 |
1.3 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
4.9 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.2 |
1.1 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.2 |
0.9 |
GO:0033688 |
regulation of osteoblast proliferation(GO:0033688) |
| 0.2 |
4.5 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.2 |
5.1 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.2 |
0.2 |
GO:0031638 |
zymogen activation(GO:0031638) |
| 0.2 |
0.4 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 |
0.4 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 |
1.0 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.2 |
0.6 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.2 |
4.2 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.2 |
0.4 |
GO:0060457 |
negative regulation of digestive system process(GO:0060457) |
| 0.2 |
0.2 |
GO:0050820 |
positive regulation of coagulation(GO:0050820) |
| 0.2 |
0.2 |
GO:0060157 |
urinary bladder development(GO:0060157) |
| 0.2 |
0.2 |
GO:0019530 |
taurine metabolic process(GO:0019530) |
| 0.2 |
4.9 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.2 |
11.1 |
GO:0090132 |
epithelial cell migration(GO:0010631) tissue migration(GO:0090130) epithelium migration(GO:0090132) |
| 0.2 |
0.2 |
GO:0019883 |
antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.2 |
0.6 |
GO:0010002 |
cardioblast differentiation(GO:0010002) |
| 0.2 |
1.4 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.2 |
3.1 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.2 |
1.7 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.2 |
0.4 |
GO:0060013 |
synaptic transmission, glycinergic(GO:0060012) righting reflex(GO:0060013) |
| 0.2 |
3.8 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.2 |
2.6 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.2 |
0.6 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 |
0.2 |
GO:0034244 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 |
0.7 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.2 |
5.7 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.2 |
0.4 |
GO:0045415 |
negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.2 |
0.9 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 |
1.8 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.2 |
1.4 |
GO:0007567 |
parturition(GO:0007567) |
| 0.2 |
0.2 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
2.8 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.2 |
0.9 |
GO:0009756 |
carbohydrate mediated signaling(GO:0009756) |
| 0.2 |
0.5 |
GO:0007184 |
SMAD protein import into nucleus(GO:0007184) |
| 0.2 |
4.2 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 |
0.3 |
GO:0045768 |
obsolete positive regulation of anti-apoptosis(GO:0045768) |
| 0.2 |
0.7 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.2 |
0.3 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 |
2.8 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.2 |
2.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.2 |
0.5 |
GO:0071347 |
cellular response to interleukin-1(GO:0071347) |
| 0.2 |
0.2 |
GO:0070244 |
negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.2 |
0.3 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.7 |
GO:0032465 |
regulation of cytokinesis(GO:0032465) |
| 0.2 |
2.0 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.2 |
0.5 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.2 |
1.7 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.2 |
0.2 |
GO:0014066 |
regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.2 |
0.7 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 |
0.7 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.2 |
0.2 |
GO:0022605 |
oogenesis stage(GO:0022605) |
| 0.2 |
0.7 |
GO:0048635 |
negative regulation of muscle organ development(GO:0048635) |
| 0.2 |
0.5 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.2 |
0.2 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.2 |
0.2 |
GO:0050685 |
regulation of mRNA 3'-end processing(GO:0031440) positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.2 |
1.3 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.2 |
1.8 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.2 |
22.4 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.2 |
0.2 |
GO:0050907 |
detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.2 |
1.3 |
GO:0045542 |
positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.2 |
0.6 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 |
0.5 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.2 |
0.3 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 |
0.8 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.2 |
0.8 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.2 |
0.2 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.2 |
1.9 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.2 |
0.8 |
GO:0006789 |
bilirubin conjugation(GO:0006789) biphenyl metabolic process(GO:0018879) biphenyl catabolic process(GO:0070980) |
| 0.2 |
1.3 |
GO:0048008 |
platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.2 |
5.8 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.2 |
0.5 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.2 |
3.3 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 |
2.3 |
GO:0006693 |
prostaglandin metabolic process(GO:0006693) |
| 0.2 |
1.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.2 |
0.8 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.2 |
0.5 |
GO:0003214 |
cardiac left ventricle morphogenesis(GO:0003214) |
| 0.2 |
0.5 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.2 |
2.3 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.2 |
0.5 |
GO:0071918 |
urea transmembrane transport(GO:0071918) |
| 0.2 |
0.2 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.2 |
0.5 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.2 |
13.9 |
GO:0008154 |
actin polymerization or depolymerization(GO:0008154) |
| 0.1 |
0.4 |
GO:0043012 |
regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.1 |
0.9 |
GO:0010766 |
negative regulation of sodium ion transport(GO:0010766) |
| 0.1 |
1.9 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.1 |
0.3 |
GO:0042159 |
lipoprotein catabolic process(GO:0042159) |
| 0.1 |
0.4 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.1 |
GO:1901998 |
antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.1 |
0.4 |
GO:0006007 |
glucose catabolic process(GO:0006007) |
| 0.1 |
0.6 |
GO:0035051 |
cardiocyte differentiation(GO:0035051) |
| 0.1 |
2.5 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.1 |
0.4 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.1 |
3.3 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.1 |
0.9 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.7 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.1 |
GO:0051590 |
positive regulation of neurotransmitter transport(GO:0051590) |
| 0.1 |
0.7 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
0.1 |
GO:0003321 |
positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.1 |
0.6 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 |
0.3 |
GO:0032236 |
obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 |
3.1 |
GO:0045995 |
regulation of embryonic development(GO:0045995) |
| 0.1 |
0.3 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.1 |
0.4 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.1 |
4.5 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 |
0.7 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 |
0.3 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.8 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.1 |
0.3 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 |
0.8 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 |
8.8 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 |
1.2 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.1 |
0.5 |
GO:0043128 |
regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 |
0.3 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.3 |
GO:1901031 |
regulation of cellular response to oxidative stress(GO:1900407) regulation of response to reactive oxygen species(GO:1901031) regulation of response to oxidative stress(GO:1902882) |
| 0.1 |
0.7 |
GO:0007351 |
blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 |
0.1 |
GO:0010269 |
response to selenium ion(GO:0010269) |
| 0.1 |
0.1 |
GO:0051155 |
positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 |
0.8 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.1 |
0.1 |
GO:0034384 |
high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 |
0.8 |
GO:0043526 |
obsolete neuroprotection(GO:0043526) |
| 0.1 |
0.1 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 |
0.6 |
GO:0010976 |
positive regulation of neuron projection development(GO:0010976) |
| 0.1 |
0.5 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.1 |
0.6 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.1 |
1.3 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 |
0.5 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
0.1 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 |
1.6 |
GO:0070193 |
synaptonemal complex organization(GO:0070193) |
| 0.1 |
2.8 |
GO:0007613 |
memory(GO:0007613) |
| 0.1 |
5.0 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.1 |
0.5 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 |
0.4 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.2 |
GO:0033144 |
negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.1 |
0.2 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.1 |
3.2 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
2.1 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 |
0.2 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.1 |
1.4 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.1 |
0.2 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.1 |
0.2 |
GO:0046322 |
negative regulation of fatty acid oxidation(GO:0046322) |
| 0.1 |
0.5 |
GO:0006029 |
proteoglycan metabolic process(GO:0006029) |
| 0.1 |
0.1 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.2 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 |
0.7 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.1 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 |
0.8 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.1 |
0.5 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.2 |
GO:0014044 |
Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 |
0.2 |
GO:0032329 |
L-serine transport(GO:0015825) serine transport(GO:0032329) |
| 0.1 |
0.4 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.1 |
2.5 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
7.6 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.1 |
0.5 |
GO:1903332 |
regulation of protein folding(GO:1903332) |
| 0.1 |
0.6 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
9.6 |
GO:0007565 |
female pregnancy(GO:0007565) |
| 0.1 |
0.1 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.6 |
GO:0031333 |
negative regulation of protein complex assembly(GO:0031333) |
| 0.1 |
1.5 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.1 |
0.3 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.1 |
0.9 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.1 |
0.4 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.1 |
GO:0002885 |
positive regulation of hypersensitivity(GO:0002885) |
| 0.1 |
0.8 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.1 |
2.0 |
GO:0030166 |
proteoglycan biosynthetic process(GO:0030166) |
| 0.1 |
0.5 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 |
0.2 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.1 |
0.2 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.3 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 |
0.1 |
GO:0000271 |
polysaccharide biosynthetic process(GO:0000271) |
| 0.1 |
2.8 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.1 |
0.4 |
GO:1902668 |
negative regulation of axon guidance(GO:1902668) |
| 0.1 |
0.5 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 |
0.7 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 |
0.1 |
GO:0060253 |
negative regulation of glial cell proliferation(GO:0060253) |
| 0.1 |
0.3 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 |
0.1 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.1 |
0.2 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 |
0.7 |
GO:0042226 |
interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 |
0.5 |
GO:0043616 |
keratinocyte proliferation(GO:0043616) |
| 0.1 |
0.3 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.5 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.3 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.1 |
0.3 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.1 |
2.8 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.1 |
0.3 |
GO:0002124 |
aggressive behavior(GO:0002118) territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.1 |
0.2 |
GO:0030858 |
positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.1 |
0.8 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.1 |
0.2 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.1 |
0.1 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
0.6 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.1 |
0.5 |
GO:0030335 |
positive regulation of cell migration(GO:0030335) positive regulation of cell motility(GO:2000147) |
| 0.1 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.1 |
0.2 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.1 |
2.3 |
GO:0015718 |
monocarboxylic acid transport(GO:0015718) |
| 0.1 |
1.9 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.1 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.1 |
0.2 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting two-sector ATPase complex assembly(GO:0070071) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
0.3 |
GO:0051102 |
DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 |
0.1 |
GO:0046618 |
drug export(GO:0046618) |
| 0.1 |
0.2 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 |
0.8 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.2 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of nucleoside metabolic process(GO:0045979) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) positive regulation of ATP metabolic process(GO:1903580) |
| 0.1 |
0.2 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 |
0.9 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.1 |
3.9 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.1 |
0.2 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.1 |
0.2 |
GO:0002902 |
regulation of B cell apoptotic process(GO:0002902) negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.5 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.1 |
0.3 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
7.4 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 |
0.9 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 |
1.1 |
GO:0061035 |
regulation of cartilage development(GO:0061035) |
| 0.1 |
0.2 |
GO:0046495 |
nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 |
0.8 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.1 |
0.1 |
GO:0015893 |
drug transport(GO:0015893) |
| 0.1 |
0.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.4 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.2 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) regulation of hair follicle development(GO:0051797) positive regulation of hair follicle development(GO:0051798) |
| 0.1 |
0.4 |
GO:0008589 |
regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 |
0.3 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.4 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.5 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.1 |
0.3 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 |
0.2 |
GO:0007144 |
female meiotic division(GO:0007143) female meiosis I(GO:0007144) |
| 0.1 |
0.1 |
GO:0033025 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 |
0.1 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 |
0.7 |
GO:0070972 |
protein targeting to ER(GO:0045047) protein localization to endoplasmic reticulum(GO:0070972) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.1 |
0.2 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.3 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.5 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.1 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 |
0.7 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.1 |
0.3 |
GO:0043441 |
acetoacetic acid biosynthetic process(GO:0043441) |
| 0.1 |
0.2 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.1 |
3.0 |
GO:0071377 |
cellular response to glucagon stimulus(GO:0071377) |
| 0.1 |
1.9 |
GO:0043547 |
positive regulation of GTPase activity(GO:0043547) |
| 0.1 |
0.3 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
3.0 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 |
0.2 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.4 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.1 |
0.3 |
GO:0001990 |
regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 |
0.8 |
GO:0032355 |
response to estradiol(GO:0032355) |
| 0.1 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
2.0 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 |
0.2 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.3 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.1 |
0.6 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.1 |
GO:2000050 |
regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.1 |
1.4 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
0.5 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 |
0.2 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.1 |
0.2 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.1 |
0.6 |
GO:0030502 |
negative regulation of bone mineralization(GO:0030502) |
| 0.1 |
0.5 |
GO:0072595 |
maintenance of protein localization in organelle(GO:0072595) |
| 0.1 |
0.1 |
GO:1902932 |
positive regulation of alcohol biosynthetic process(GO:1902932) |
| 0.1 |
0.1 |
GO:0046717 |
acid secretion(GO:0046717) |
| 0.1 |
0.6 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
1.5 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 |
5.0 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.1 |
1.3 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 |
1.2 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.1 |
0.1 |
GO:0051934 |
dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.1 |
0.3 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 |
0.9 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.1 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.1 |
GO:0030728 |
ovulation(GO:0030728) |
| 0.1 |
6.9 |
GO:0008544 |
epidermis development(GO:0008544) |
| 0.1 |
0.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.4 |
GO:0090280 |
positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) positive regulation of calcium ion import(GO:0090280) |
| 0.1 |
0.1 |
GO:0034650 |
cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.1 |
0.2 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.1 |
GO:0046856 |
phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 |
0.4 |
GO:0032786 |
positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 |
0.2 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 |
0.2 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.9 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.1 |
GO:0030324 |
lung development(GO:0030324) |
| 0.1 |
0.2 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.1 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.1 |
0.3 |
GO:0048016 |
inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 |
0.1 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.1 |
0.1 |
GO:0009698 |
phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.1 |
0.1 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.1 |
0.1 |
GO:0050666 |
regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 |
0.1 |
GO:2001021 |
negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 |
0.4 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
1.2 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 |
0.4 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 |
0.1 |
GO:0014020 |
primary neural tube formation(GO:0014020) |
| 0.1 |
1.0 |
GO:0006665 |
sphingolipid metabolic process(GO:0006665) |
| 0.1 |
0.1 |
GO:0032506 |
barrier septum assembly(GO:0000917) cytokinetic process(GO:0032506) cell septum assembly(GO:0090529) mitotic cytokinetic process(GO:1902410) |
| 0.1 |
0.1 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 |
1.2 |
GO:0030336 |
negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.1 |
0.4 |
GO:0035308 |
negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 |
0.2 |
GO:0009313 |
oligosaccharide catabolic process(GO:0009313) |
| 0.1 |
0.4 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.7 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.1 |
0.8 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.1 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.2 |
GO:0010889 |
regulation of sequestering of triglyceride(GO:0010889) |
| 0.1 |
0.9 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 |
2.8 |
GO:0071772 |
BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.1 |
0.4 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.8 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 |
0.9 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.1 |
21.4 |
GO:0048858 |
cell projection morphogenesis(GO:0048858) |
| 0.1 |
0.3 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 |
0.1 |
GO:0048048 |
embryonic eye morphogenesis(GO:0048048) |
| 0.1 |
0.2 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 |
0.2 |
GO:0017014 |
protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 |
0.1 |
GO:0044818 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) mitotic G2/M transition checkpoint(GO:0044818) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 |
0.2 |
GO:0051140 |
regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 |
0.6 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.1 |
0.5 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 |
5.8 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.1 |
0.2 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 |
0.7 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.2 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
1.0 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.1 |
0.7 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
1.1 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 |
0.2 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 |
0.2 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
0.1 |
GO:0090042 |
tubulin deacetylation(GO:0090042) |
| 0.1 |
0.3 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.1 |
0.2 |
GO:0021548 |
pons development(GO:0021548) |
| 0.1 |
0.4 |
GO:0031345 |
negative regulation of cell projection organization(GO:0031345) |
| 0.1 |
0.2 |
GO:0017004 |
cytochrome complex assembly(GO:0017004) |
| 0.1 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.1 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.1 |
0.2 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.1 |
0.1 |
GO:0032026 |
response to magnesium ion(GO:0032026) |
| 0.1 |
0.1 |
GO:0046626 |
regulation of insulin receptor signaling pathway(GO:0046626) regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.0 |
0.3 |
GO:0034374 |
low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 |
0.1 |
GO:0051552 |
flavone metabolic process(GO:0051552) |
| 0.0 |
0.1 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.0 |
0.2 |
GO:0002062 |
chondrocyte differentiation(GO:0002062) |
| 0.0 |
2.3 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.1 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
1.6 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.2 |
GO:0018101 |
protein citrullination(GO:0018101) citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.0 |
GO:0043046 |
DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 |
0.2 |
GO:0009629 |
response to gravity(GO:0009629) |
| 0.0 |
0.1 |
GO:0071109 |
striatum development(GO:0021756) superior temporal gyrus development(GO:0071109) |
| 0.0 |
0.1 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 |
0.9 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.8 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
5.7 |
GO:0071466 |
xenobiotic metabolic process(GO:0006805) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 |
1.4 |
GO:0002576 |
platelet degranulation(GO:0002576) |
| 0.0 |
0.1 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.4 |
GO:0032507 |
maintenance of protein location in cell(GO:0032507) |
| 0.0 |
2.3 |
GO:0034249 |
negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 |
0.0 |
GO:0051322 |
anaphase(GO:0051322) |
| 0.0 |
1.6 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.0 |
0.1 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 |
0.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.7 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.3 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.0 |
GO:0032847 |
regulation of cellular pH reduction(GO:0032847) |
| 0.0 |
0.6 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.8 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 |
0.2 |
GO:0002517 |
T cell tolerance induction(GO:0002517) regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 |
0.1 |
GO:0001547 |
antral ovarian follicle growth(GO:0001547) |
| 0.0 |
1.7 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.3 |
GO:0001933 |
negative regulation of protein phosphorylation(GO:0001933) |
| 0.0 |
0.1 |
GO:0048003 |
antigen processing and presentation via MHC class Ib(GO:0002475) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 |
0.0 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 |
0.0 |
GO:0002507 |
tolerance induction(GO:0002507) |
| 0.0 |
0.6 |
GO:0030282 |
bone mineralization(GO:0030282) |
| 0.0 |
0.2 |
GO:0014033 |
neural crest cell differentiation(GO:0014033) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.1 |
GO:0006423 |
cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 |
0.1 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.0 |
GO:0031952 |
regulation of protein autophosphorylation(GO:0031952) |
| 0.0 |
0.1 |
GO:1903510 |
mucopolysaccharide metabolic process(GO:1903510) |
| 0.0 |
0.0 |
GO:0042523 |
regulation of tyrosine phosphorylation of Stat5 protein(GO:0042522) positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 |
0.0 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.1 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.0 |
0.3 |
GO:0030048 |
actin filament-based movement(GO:0030048) |
| 0.0 |
0.1 |
GO:0071375 |
cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 |
1.7 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.1 |
GO:0048588 |
developmental cell growth(GO:0048588) |
| 0.0 |
0.1 |
GO:0070243 |
regulation of thymocyte apoptotic process(GO:0070243) |
| 0.0 |
0.1 |
GO:0045936 |
negative regulation of phosphorus metabolic process(GO:0010563) negative regulation of phosphate metabolic process(GO:0045936) |
| 0.0 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 |
0.1 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0006978 |
DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 |
0.0 |
GO:0048486 |
parasympathetic nervous system development(GO:0048486) |
| 0.0 |
0.2 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 |
0.2 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.0 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.1 |
GO:0014896 |
muscle hypertrophy(GO:0014896) |
| 0.0 |
0.2 |
GO:0045766 |
positive regulation of angiogenesis(GO:0045766) |
| 0.0 |
0.2 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.0 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.0 |
0.4 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 |
0.1 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.0 |
2.1 |
GO:0034329 |
cell junction assembly(GO:0034329) |
| 0.0 |
3.4 |
GO:0006814 |
sodium ion transport(GO:0006814) |
| 0.0 |
1.6 |
GO:0031589 |
cell-substrate adhesion(GO:0031589) |
| 0.0 |
0.0 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.3 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.1 |
GO:0046321 |
positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 |
0.2 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
1.9 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.0 |
GO:1902624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 |
0.3 |
GO:0046464 |
triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 |
0.1 |
GO:0015811 |
sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 |
0.4 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.0 |
0.1 |
GO:0006531 |
aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 |
0.1 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.0 |
0.2 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 |
0.1 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.0 |
0.1 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.2 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.0 |
0.1 |
GO:0048485 |
sympathetic nervous system development(GO:0048485) |
| 0.0 |
0.1 |
GO:0042311 |
vasodilation(GO:0042311) |
| 0.0 |
0.0 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.0 |
0.4 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
2.1 |
GO:0006487 |
protein N-linked glycosylation(GO:0006487) |
| 0.0 |
0.3 |
GO:0035303 |
regulation of dephosphorylation(GO:0035303) |
| 0.0 |
0.1 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.1 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
2.1 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.0 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.2 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.1 |
GO:0045350 |
interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 |
0.1 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.0 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.9 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.0 |
0.0 |
GO:0042510 |
regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) |
| 0.0 |
0.1 |
GO:0045598 |
regulation of fat cell differentiation(GO:0045598) |
| 0.0 |
0.1 |
GO:0045713 |
low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 |
0.0 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 |
0.0 |
GO:0001963 |
synaptic transmission, dopaminergic(GO:0001963) |
| 0.0 |
0.1 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
0.1 |
GO:0015722 |
canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 |
0.0 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
2.3 |
GO:0052547 |
regulation of peptidase activity(GO:0052547) |
| 0.0 |
0.1 |
GO:0071295 |
cellular response to vitamin(GO:0071295) |
| 0.0 |
0.5 |
GO:0007623 |
circadian rhythm(GO:0007623) |
| 0.0 |
0.1 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.0 |
0.1 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.2 |
GO:0042752 |
regulation of circadian rhythm(GO:0042752) |
| 0.0 |
0.0 |
GO:0014721 |
voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 |
0.0 |
GO:0072676 |
lymphocyte chemotaxis(GO:0048247) lymphocyte migration(GO:0072676) |
| 0.0 |
0.0 |
GO:0009438 |
methylglyoxal metabolic process(GO:0009438) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.0 |
GO:0050808 |
synapse organization(GO:0050808) |
| 0.0 |
0.4 |
GO:0045444 |
fat cell differentiation(GO:0045444) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.5 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.1 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
0.2 |
GO:0046320 |
regulation of fatty acid oxidation(GO:0046320) |
| 0.0 |
0.1 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.1 |
GO:0010390 |
histone monoubiquitination(GO:0010390) |
| 0.0 |
0.1 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.0 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 |
0.2 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.0 |
GO:0033158 |
regulation of protein import into nucleus, translocation(GO:0033158) |
| 0.0 |
0.7 |
GO:0007586 |
digestion(GO:0007586) |
| 0.0 |
0.2 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.5 |
GO:0007338 |
single fertilization(GO:0007338) |
| 0.0 |
0.0 |
GO:1902600 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 |
0.1 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.0 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.0 |
GO:0051324 |
prophase(GO:0051324) |
| 0.0 |
0.0 |
GO:0007468 |
regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 |
0.0 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.0 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.0 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 |
0.0 |
GO:0072332 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.0 |
GO:0043588 |
skin development(GO:0043588) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |