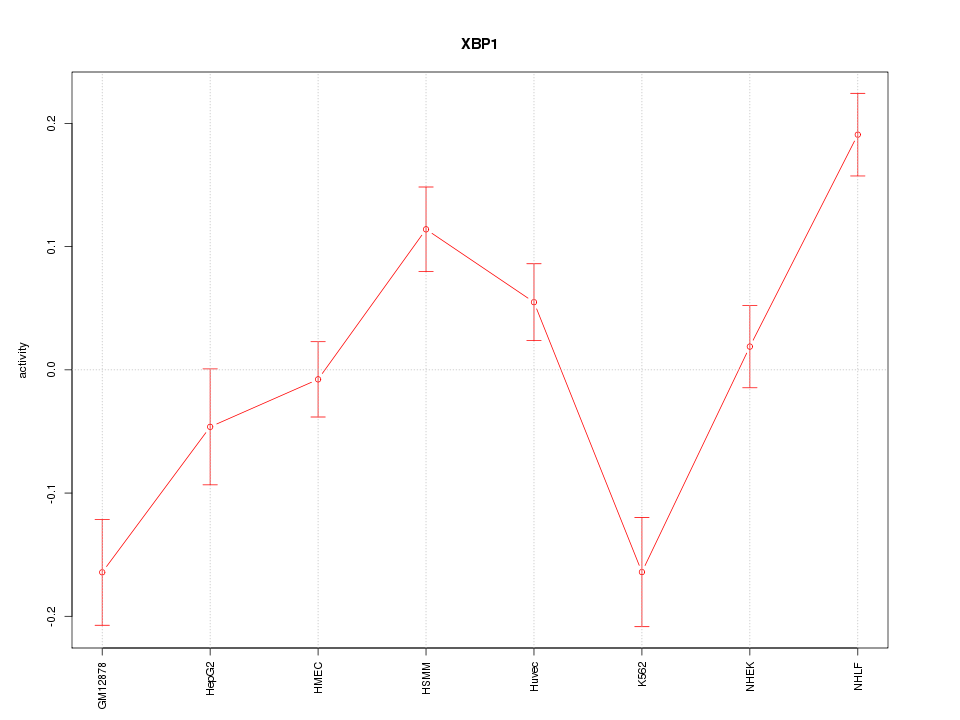
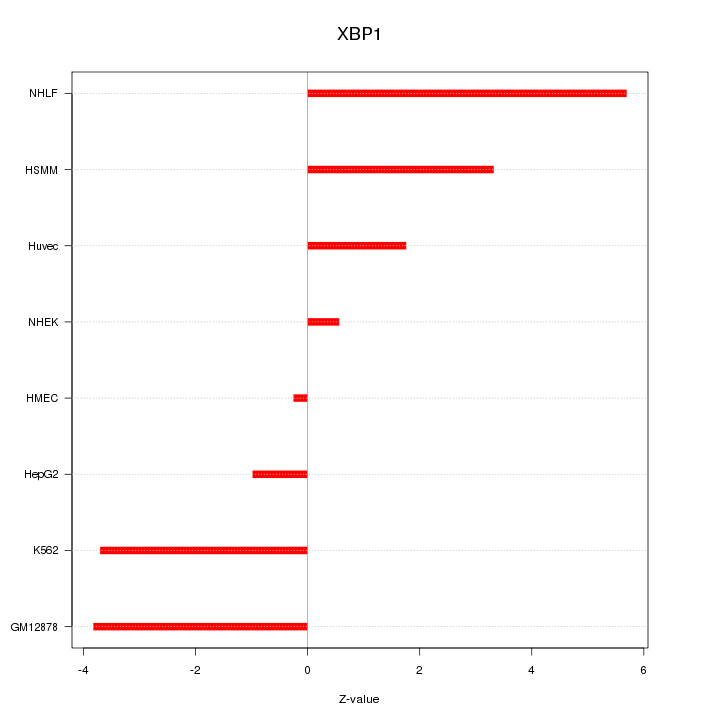
Motif ID: XBP1
Z-value: 3.087
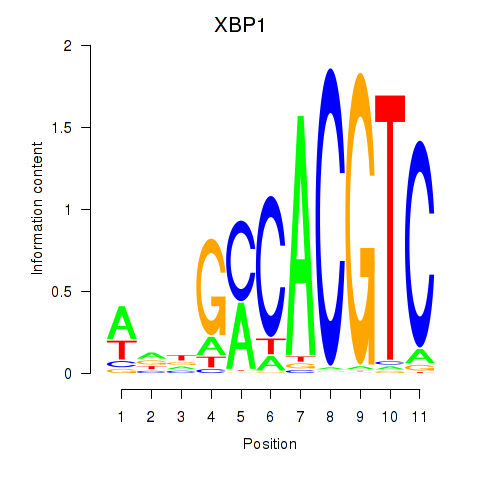
Transcription factors associated with XBP1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| XBP1 | ENSG00000100219.12 | XBP1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 2.5 | 7.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.5 | 5.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.4 | 5.4 | GO:0033197 | response to vitamin E(GO:0033197) |
| 1.1 | 3.3 | GO:0060574 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 1.0 | 3.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.8 | 2.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 6.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.8 | 8.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.7 | 4.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.6 | 1.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.5 | 1.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.5 | 1.9 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 3.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 1.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 1.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.3 | 1.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 1.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 3.5 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.2 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 2.4 | GO:0070972 | protein targeting to ER(GO:0045047) protein localization to endoplasmic reticulum(GO:0070972) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.2 | 9.8 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.2 | 1.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.2 | 3.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 2.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 7.7 | GO:0008064 | regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 | 2.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 2.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.1 | 2.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 1.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 4.0 | GO:0006914 | autophagy(GO:0006914) |
| 0.1 | 1.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 7.0 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.9 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 1.2 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.5 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.3 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.5 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.1 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 2.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 | 2.3 | GO:0000236 | mitotic prometaphase(GO:0000236) |
| 0.0 | 5.1 | GO:0043623 | cellular protein complex assembly(GO:0043623) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 2.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 1.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 21.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 7.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 1.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.5 | 4.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.4 | 7.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 1.4 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 3.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 5.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 6.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 7.2 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0019718 | obsolete rough microsome(GO:0019718) |
| 0.0 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 1.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 8.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 6.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 22.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.9 | 21.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 8.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.5 | 1.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.5 | 13.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.5 | 1.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 6.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.4 | 3.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 1.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 1.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 1.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 5.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.9 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 1.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) nucleotide diphosphatase activity(GO:0004551) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 11.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 9.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.2 | 5.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 7.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.9 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |