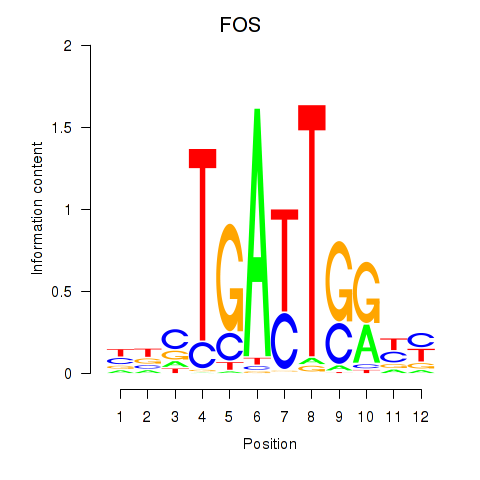
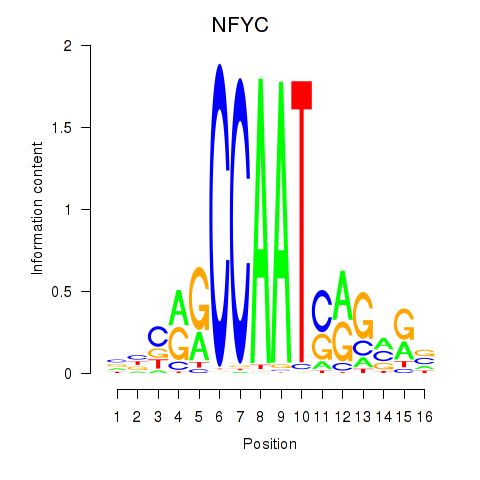
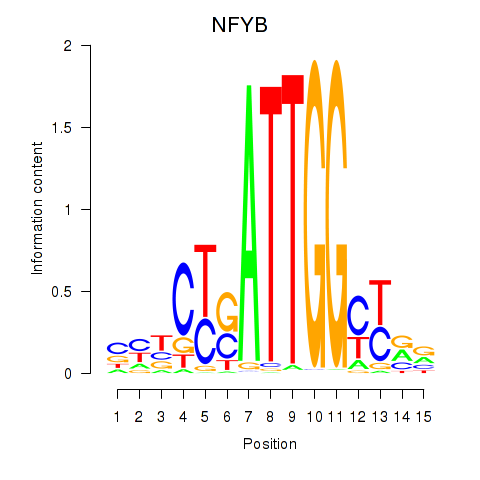
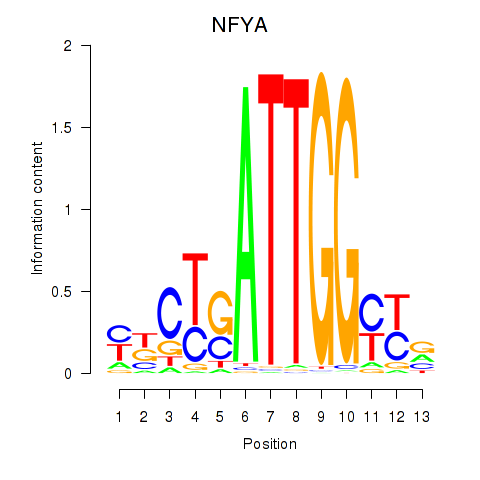
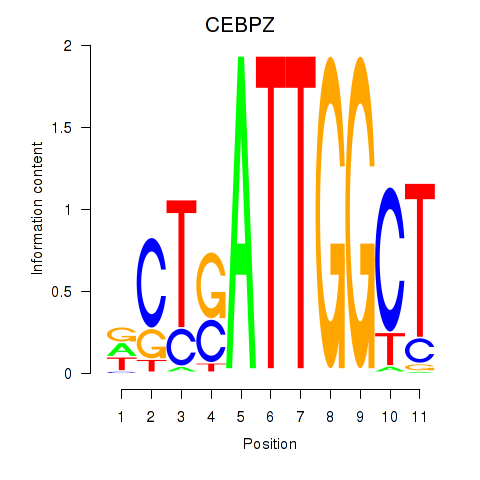
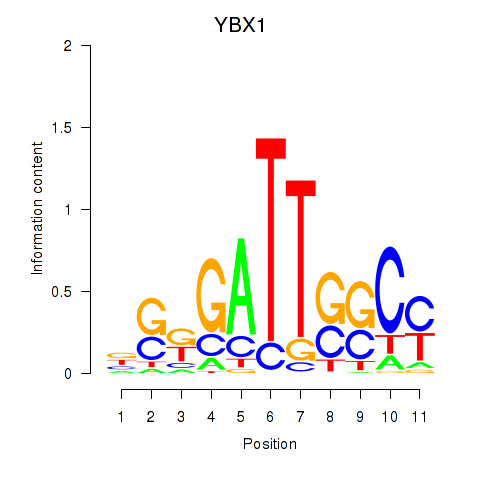
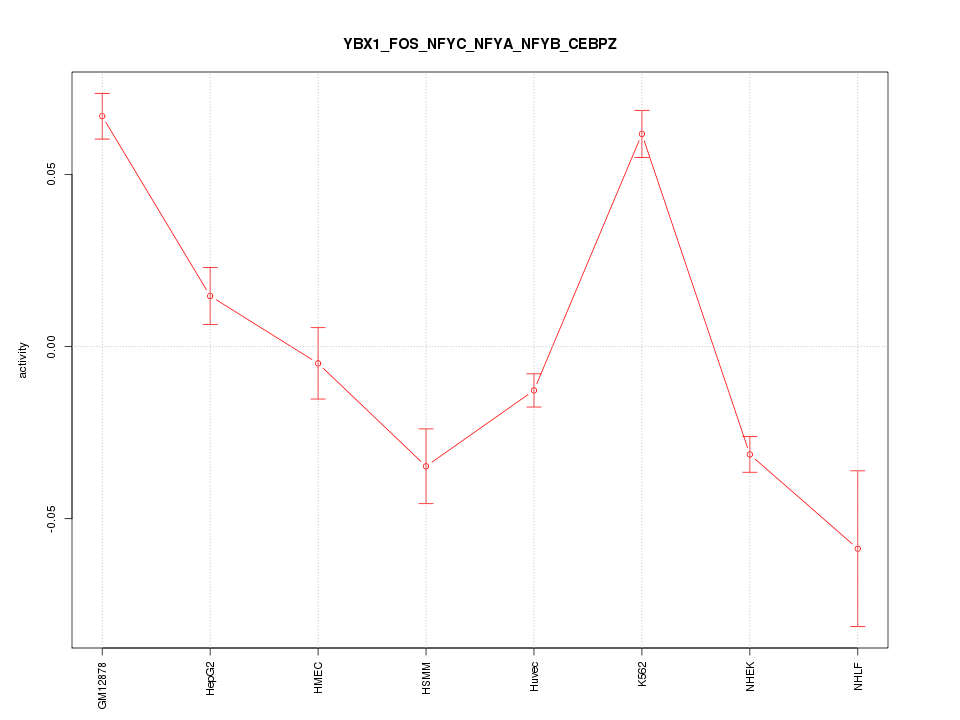
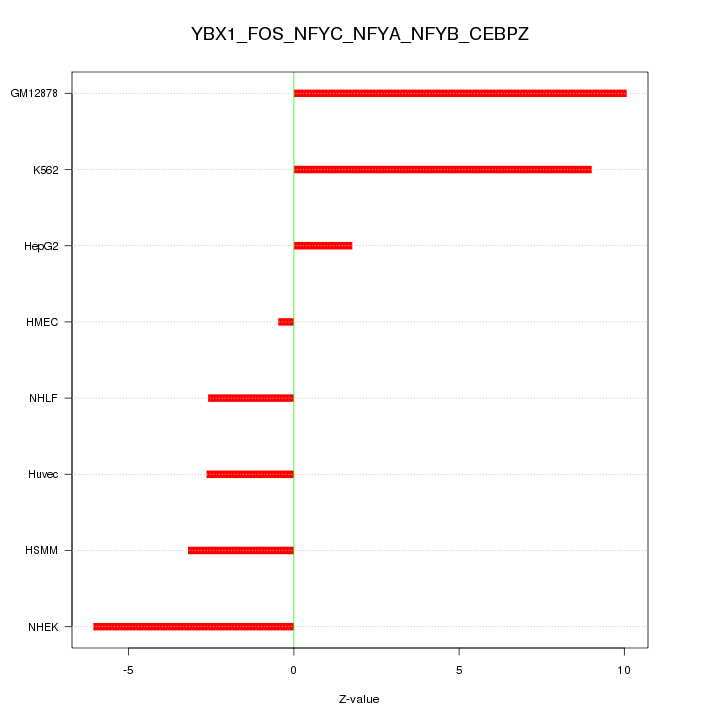
| 3.1 |
9.2 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 2.5 |
15.0 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 2.5 |
4.9 |
GO:0000216 |
obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 2.2 |
6.7 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 2.0 |
7.9 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 2.0 |
7.8 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 1.9 |
5.8 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 1.9 |
7.5 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 1.9 |
7.5 |
GO:0045950 |
negative regulation of mitotic recombination(GO:0045950) |
| 1.9 |
3.8 |
GO:0071459 |
protein localization to chromosome, centromeric region(GO:0071459) |
| 1.8 |
5.4 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 1.7 |
5.0 |
GO:0010041 |
response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 1.7 |
6.7 |
GO:0006768 |
biotin metabolic process(GO:0006768) |
| 1.6 |
3.3 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 1.6 |
4.8 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 1.6 |
6.4 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 1.6 |
4.7 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 1.5 |
4.5 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 1.5 |
16.0 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 1.4 |
7.2 |
GO:0000089 |
mitotic metaphase(GO:0000089) |
| 1.4 |
4.2 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 1.4 |
9.7 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 1.4 |
9.6 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 1.4 |
11.0 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 1.4 |
5.5 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 1.3 |
6.6 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 1.2 |
3.7 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.2 |
8.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 1.2 |
5.9 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.2 |
3.5 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 1.2 |
3.5 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 1.2 |
1.2 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 1.2 |
4.6 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 1.1 |
4.6 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 1.1 |
5.5 |
GO:0006189 |
'de novo' IMP biosynthetic process(GO:0006189) |
| 1.0 |
5.2 |
GO:0044818 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) mitotic G2/M transition checkpoint(GO:0044818) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 1.0 |
6.2 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 1.0 |
3.0 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 1.0 |
4.9 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 1.0 |
2.9 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 1.0 |
2.9 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 1.0 |
7.6 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.9 |
7.5 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.9 |
5.6 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.9 |
20.3 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.9 |
2.8 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.9 |
2.7 |
GO:2000644 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.9 |
78.2 |
GO:0031497 |
chromatin assembly(GO:0031497) |
| 0.9 |
3.6 |
GO:0032070 |
regulation of deoxyribonuclease activity(GO:0032070) |
| 0.9 |
5.3 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.9 |
0.9 |
GO:0090206 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.9 |
2.6 |
GO:0034635 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 |
3.4 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.8 |
1.7 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.8 |
3.3 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.8 |
2.5 |
GO:0034626 |
fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 |
5.8 |
GO:0042797 |
5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.8 |
2.5 |
GO:0000019 |
regulation of mitotic recombination(GO:0000019) |
| 0.8 |
5.8 |
GO:0040001 |
establishment of mitotic spindle localization(GO:0040001) |
| 0.8 |
4.8 |
GO:0051383 |
centromere complex assembly(GO:0034508) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.8 |
5.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.8 |
0.8 |
GO:0043523 |
regulation of neuron apoptotic process(GO:0043523) regulation of neuron death(GO:1901214) |
| 0.8 |
3.1 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.8 |
6.8 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.8 |
0.8 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) negative regulation of stem cell differentiation(GO:2000737) |
| 0.7 |
2.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.7 |
2.9 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.7 |
2.7 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.7 |
4.0 |
GO:0051299 |
centrosome separation(GO:0051299) |
| 0.7 |
2.0 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.7 |
2.0 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.6 |
1.3 |
GO:0001832 |
blastocyst growth(GO:0001832) inner cell mass cell proliferation(GO:0001833) |
| 0.6 |
5.5 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.6 |
1.2 |
GO:0051892 |
negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.6 |
7.9 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.6 |
1.8 |
GO:0070813 |
cysteine biosynthetic process(GO:0019344) hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.6 |
1.7 |
GO:0097237 |
cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.6 |
3.5 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 |
1.1 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.6 |
1.7 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.6 |
2.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.6 |
1.7 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.5 |
2.7 |
GO:0045953 |
negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.5 |
2.7 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.5 |
2.1 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.5 |
16.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.5 |
2.1 |
GO:0006565 |
L-serine catabolic process(GO:0006565) |
| 0.5 |
2.1 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.5 |
1.5 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.5 |
1.5 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.5 |
2.5 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.5 |
4.5 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.5 |
3.5 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.5 |
3.0 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.5 |
1.5 |
GO:0051231 |
mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.5 |
3.4 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.5 |
1.5 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.5 |
0.5 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 |
2.4 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.5 |
42.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.5 |
3.2 |
GO:0045824 |
negative regulation of innate immune response(GO:0045824) |
| 0.4 |
1.3 |
GO:0046102 |
nicotinamide riboside catabolic process(GO:0006738) inosine metabolic process(GO:0046102) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.4 |
1.8 |
GO:0072180 |
mesonephric duct morphogenesis(GO:0072180) |
| 0.4 |
1.3 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.4 |
1.3 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
4.6 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.4 |
1.2 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.4 |
3.6 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.4 |
1.6 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.4 |
1.5 |
GO:0070933 |
histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.4 |
7.3 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.4 |
1.9 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.4 |
1.1 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.4 |
1.1 |
GO:0019521 |
aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.4 |
0.7 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.4 |
0.4 |
GO:0043269 |
regulation of ion transport(GO:0043269) |
| 0.4 |
2.8 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.3 |
0.7 |
GO:0019322 |
pentose biosynthetic process(GO:0019322) |
| 0.3 |
1.7 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.3 |
1.0 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 |
2.1 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 |
1.0 |
GO:0060152 |
peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.3 |
1.7 |
GO:0051570 |
regulation of histone H3-K9 methylation(GO:0051570) |
| 0.3 |
1.0 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.3 |
1.0 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.3 |
1.0 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.3 |
1.9 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.3 |
2.9 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.3 |
3.9 |
GO:0055069 |
cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.3 |
1.3 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.3 |
1.6 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 |
0.9 |
GO:1902745 |
positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.3 |
1.8 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.3 |
0.9 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 |
0.9 |
GO:1904742 |
regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.3 |
1.2 |
GO:0046778 |
modification by virus of host mRNA processing(GO:0046778) |
| 0.3 |
0.9 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.3 |
6.7 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.3 |
2.0 |
GO:0045007 |
depurination(GO:0045007) |
| 0.3 |
2.0 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.3 |
3.7 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.3 |
1.1 |
GO:0009103 |
lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.3 |
4.4 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.3 |
0.5 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.3 |
0.8 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.3 |
0.8 |
GO:0050685 |
positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.3 |
1.3 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.3 |
2.0 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 |
1.7 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.2 |
3.0 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 |
13.7 |
GO:0008630 |
intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.2 |
1.0 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.2 |
2.2 |
GO:0009744 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 |
0.5 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.2 |
1.9 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.2 |
0.5 |
GO:0030578 |
nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.2 |
0.5 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.2 |
0.9 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.2 |
0.7 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 |
1.2 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.2 |
3.0 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 |
1.4 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 |
0.7 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.2 |
0.4 |
GO:2000309 |
tumor necrosis factor (ligand) superfamily member 11 production(GO:0072535) regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000307) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.2 |
0.9 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.2 |
0.9 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 |
0.9 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.2 |
2.2 |
GO:0034644 |
cellular response to UV(GO:0034644) |
| 0.2 |
0.9 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
1.9 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.2 |
0.9 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 |
0.9 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
1.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 |
1.3 |
GO:0060676 |
ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.2 |
1.3 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
4.1 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.2 |
1.4 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.2 |
0.4 |
GO:0045992 |
negative regulation of embryonic development(GO:0045992) |
| 0.2 |
4.7 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.2 |
4.9 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 |
2.0 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.2 |
5.4 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 |
0.8 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.2 |
1.6 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 |
1.2 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 |
1.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 |
1.6 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 |
1.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 |
0.4 |
GO:0007231 |
osmosensory signaling pathway(GO:0007231) |
| 0.2 |
1.5 |
GO:0051005 |
negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 |
1.0 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.2 |
1.7 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.2 |
1.1 |
GO:0046545 |
development of primary female sexual characteristics(GO:0046545) |
| 0.2 |
0.6 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.2 |
0.4 |
GO:0031054 |
pre-miRNA processing(GO:0031054) |
| 0.2 |
2.0 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 |
5.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.2 |
0.7 |
GO:0031047 |
gene silencing by RNA(GO:0031047) |
| 0.2 |
1.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 |
2.1 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.2 |
0.5 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.2 |
0.5 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.2 |
1.0 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.2 |
2.3 |
GO:0050718 |
positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 |
0.2 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 |
0.7 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 |
0.5 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.2 |
0.5 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.2 |
2.9 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.2 |
0.3 |
GO:0002268 |
follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 |
0.7 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 |
0.5 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.2 |
2.0 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.2 |
0.7 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 |
0.3 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
0.6 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 |
0.2 |
GO:0072201 |
negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.2 |
0.8 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.2 |
0.5 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.2 |
1.3 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.2 |
0.5 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.2 |
0.5 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.2 |
0.3 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.2 |
6.0 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 |
0.6 |
GO:0006408 |
snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.2 |
0.8 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
0.8 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.1 |
0.4 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 |
2.7 |
GO:0010862 |
positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 |
0.6 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
0.4 |
GO:0007051 |
spindle organization(GO:0007051) |
| 0.1 |
0.4 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.1 |
1.3 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 |
0.7 |
GO:0031657 |
regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.1 |
0.9 |
GO:0050999 |
regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.1 |
0.4 |
GO:0072207 |
metanephric tubule development(GO:0072170) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.1 |
0.4 |
GO:0007090 |
obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.1 |
1.4 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
0.3 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.1 |
6.9 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.5 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.1 |
0.4 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 |
0.7 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 |
0.7 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
3.9 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.1 |
0.9 |
GO:0010737 |
protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) |
| 0.1 |
0.5 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.1 |
2.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 |
0.7 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
3.2 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 |
2.1 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.1 |
6.9 |
GO:0042632 |
cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 |
0.6 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
5.9 |
GO:0016571 |
histone methylation(GO:0016571) |
| 0.1 |
21.2 |
GO:0044843 |
G1/S transition of mitotic cell cycle(GO:0000082) cell cycle G1/S phase transition(GO:0044843) |
| 0.1 |
1.0 |
GO:0001554 |
luteolysis(GO:0001554) |
| 0.1 |
0.5 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.4 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.1 |
1.4 |
GO:0030001 |
metal ion transport(GO:0030001) |
| 0.1 |
0.6 |
GO:0046323 |
glucose import(GO:0046323) |
| 0.1 |
0.4 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 |
0.6 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 |
0.8 |
GO:0006200 |
obsolete ATP catabolic process(GO:0006200) |
| 0.1 |
0.2 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.1 |
0.6 |
GO:0035090 |
maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) |
| 0.1 |
0.8 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 |
0.3 |
GO:0090009 |
primitive streak formation(GO:0090009) |
| 0.1 |
1.4 |
GO:0042987 |
amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 |
0.3 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
1.4 |
GO:0043097 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 |
0.8 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.2 |
GO:0009750 |
response to fructose(GO:0009750) |
| 0.1 |
1.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.1 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 |
1.2 |
GO:0001947 |
heart looping(GO:0001947) embryonic heart tube morphogenesis(GO:0003143) determination of heart left/right asymmetry(GO:0061371) |
| 0.1 |
1.4 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.1 |
0.5 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.1 |
0.3 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.1 |
0.5 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
1.2 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.4 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.6 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
0.4 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.1 |
2.1 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 |
0.4 |
GO:0071480 |
smooth muscle adaptation(GO:0014805) cellular response to gamma radiation(GO:0071480) |
| 0.1 |
0.5 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 |
0.1 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.1 |
3.8 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.1 |
0.8 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 |
5.3 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.1 |
6.2 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 |
1.4 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.5 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) |
| 0.1 |
0.4 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
1.0 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
0.4 |
GO:0051216 |
cartilage development(GO:0051216) |
| 0.1 |
0.3 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.1 |
1.9 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 |
0.3 |
GO:0016445 |
somatic diversification of immunoglobulins(GO:0016445) |
| 0.1 |
0.3 |
GO:0043281 |
regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.1 |
18.0 |
GO:0000375 |
RNA splicing, via transesterification reactions(GO:0000375) |
| 0.1 |
1.5 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 |
1.5 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.1 |
3.7 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.1 |
0.3 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 |
0.2 |
GO:0046732 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by virus of host immune response(GO:0075528) |
| 0.1 |
0.5 |
GO:0090342 |
regulation of cell aging(GO:0090342) |
| 0.1 |
8.4 |
GO:0007018 |
microtubule-based movement(GO:0007018) |
| 0.1 |
0.3 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
5.4 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 |
0.7 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
1.0 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 |
4.3 |
GO:0010827 |
regulation of glucose transport(GO:0010827) |
| 0.1 |
0.4 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
0.1 |
GO:0031622 |
regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) |
| 0.1 |
2.2 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 |
1.0 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.1 |
0.6 |
GO:0048665 |
neuron fate specification(GO:0048665) |
| 0.1 |
0.1 |
GO:0000288 |
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 |
0.8 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.1 |
0.2 |
GO:0016075 |
rRNA catabolic process(GO:0016075) ncRNA catabolic process(GO:0034661) |
| 0.1 |
0.9 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
4.9 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.1 |
0.5 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 |
1.0 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.6 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.1 |
0.3 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 |
0.3 |
GO:0051026 |
chiasma assembly(GO:0051026) |
| 0.1 |
9.0 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.1 |
0.3 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.1 |
0.6 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.1 |
0.5 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.1 |
0.2 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
1.6 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.4 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 |
0.4 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.3 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.1 |
0.5 |
GO:0016073 |
snRNA metabolic process(GO:0016073) snRNA processing(GO:0016180) |
| 0.1 |
1.7 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.1 |
0.2 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 |
1.8 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 |
0.3 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.1 |
0.4 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 |
0.8 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 |
0.8 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.1 |
GO:0060746 |
maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 |
0.8 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 |
0.3 |
GO:0051452 |
intracellular pH reduction(GO:0051452) |
| 0.1 |
0.7 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
0.4 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.1 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 |
0.4 |
GO:0016114 |
terpenoid biosynthetic process(GO:0016114) |
| 0.0 |
0.8 |
GO:0042438 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.9 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.0 |
0.1 |
GO:0006531 |
aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.3 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.0 |
0.3 |
GO:0001906 |
cell killing(GO:0001906) |
| 0.0 |
0.2 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.2 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.6 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.8 |
GO:0001836 |
release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 |
0.2 |
GO:0050865 |
regulation of leukocyte activation(GO:0002694) regulation of cell activation(GO:0050865) |
| 0.0 |
0.7 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
1.0 |
GO:0051789 |
obsolete response to protein(GO:0051789) |
| 0.0 |
1.0 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0031952 |
regulation of protein autophosphorylation(GO:0031952) positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 |
1.4 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.0 |
0.3 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 |
0.2 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
1.0 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.7 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.6 |
GO:0006305 |
DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0032581 |
ER-dependent peroxisome organization(GO:0032581) |
| 0.0 |
0.1 |
GO:0072012 |
paraxial mesoderm formation(GO:0048341) renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) glomerular mesangium development(GO:0072109) |
| 0.0 |
0.9 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.8 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
4.2 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.1 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
1.0 |
GO:0051899 |
membrane depolarization(GO:0051899) |
| 0.0 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.6 |
GO:0021879 |
forebrain neuron differentiation(GO:0021879) |
| 0.0 |
0.2 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
2.0 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
1.1 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.3 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.1 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.0 |
4.8 |
GO:0006397 |
mRNA processing(GO:0006397) |
| 0.0 |
0.3 |
GO:0072348 |
sulfur compound transport(GO:0072348) |
| 0.0 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.0 |
1.0 |
GO:0032355 |
response to estradiol(GO:0032355) |
| 0.0 |
0.1 |
GO:0060157 |
urinary bladder development(GO:0060157) |
| 0.0 |
0.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.0 |
0.7 |
GO:0051341 |
regulation of oxidoreductase activity(GO:0051341) |
| 0.0 |
0.3 |
GO:0042738 |
exogenous drug catabolic process(GO:0042738) |
| 0.0 |
0.1 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.3 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.1 |
GO:0006739 |
pentose-phosphate shunt(GO:0006098) NADP metabolic process(GO:0006739) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.3 |
GO:0009411 |
response to UV(GO:0009411) |
| 0.0 |
0.1 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0098781 |
rRNA transcription(GO:0009303) ncRNA transcription(GO:0098781) |
| 0.0 |
1.3 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
0.8 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.0 |
0.6 |
GO:0006401 |
RNA catabolic process(GO:0006401) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.1 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.9 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.2 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.1 |
GO:0044091 |
peroxisome membrane biogenesis(GO:0016557) membrane biogenesis(GO:0044091) |
| 0.0 |
0.1 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 |
1.6 |
GO:0006366 |
transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 |
0.2 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
0.2 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 |
0.0 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
1.1 |
GO:0002062 |
chondrocyte differentiation(GO:0002062) |
| 0.0 |
2.5 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.3 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.2 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.4 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.2 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.3 |
GO:0006323 |
DNA packaging(GO:0006323) |
| 0.0 |
0.3 |
GO:0006302 |
double-strand break repair(GO:0006302) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 |
0.1 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 |
0.1 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 |
0.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0060178 |
exocyst assembly(GO:0001927) regulation of exocyst assembly(GO:0001928) exocyst localization(GO:0051601) regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 |
3.2 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 |
0.6 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.5 |
GO:0006695 |
cholesterol biosynthetic process(GO:0006695) |
| 0.0 |
0.2 |
GO:0001702 |
gastrulation with mouth forming second(GO:0001702) |
| 0.0 |
0.1 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.3 |
GO:0045646 |
regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 |
0.3 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 |
0.9 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
1.1 |
GO:0006310 |
DNA recombination(GO:0006310) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
1.2 |
GO:0006644 |
phospholipid metabolic process(GO:0006644) |
| 0.0 |
0.1 |
GO:0014820 |
tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 |
0.2 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.7 |
GO:0051170 |
nuclear import(GO:0051170) |
| 0.0 |
0.1 |
GO:0006266 |
DNA ligation(GO:0006266) |
| 0.0 |
0.2 |
GO:0043525 |
positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.0 |
GO:0021781 |
glial cell fate commitment(GO:0021781) |
| 0.0 |
0.9 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.2 |
GO:0045333 |
cellular respiration(GO:0045333) |
| 0.0 |
0.2 |
GO:0007215 |
glutamate receptor signaling pathway(GO:0007215) |
| 0.0 |
0.2 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.0 |
0.2 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 |
0.5 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
1.0 |
GO:0008016 |
regulation of heart contraction(GO:0008016) |
| 0.0 |
0.3 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.4 |
GO:0010952 |
positive regulation of peptidase activity(GO:0010952) |
| 0.0 |
0.0 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
21.9 |
GO:1903506 |
regulation of transcription, DNA-templated(GO:0006355) regulation of nucleic acid-templated transcription(GO:1903506) |
| 0.0 |
0.2 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.2 |
GO:0046330 |
positive regulation of JNK cascade(GO:0046330) |
| 0.0 |
0.1 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.1 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
1.9 |
GO:0006869 |
lipid transport(GO:0006869) |
| 0.0 |
0.1 |
GO:0042269 |
regulation of natural killer cell mediated immunity(GO:0002715) regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 |
0.1 |
GO:0051932 |
synaptic transmission, GABAergic(GO:0051932) |