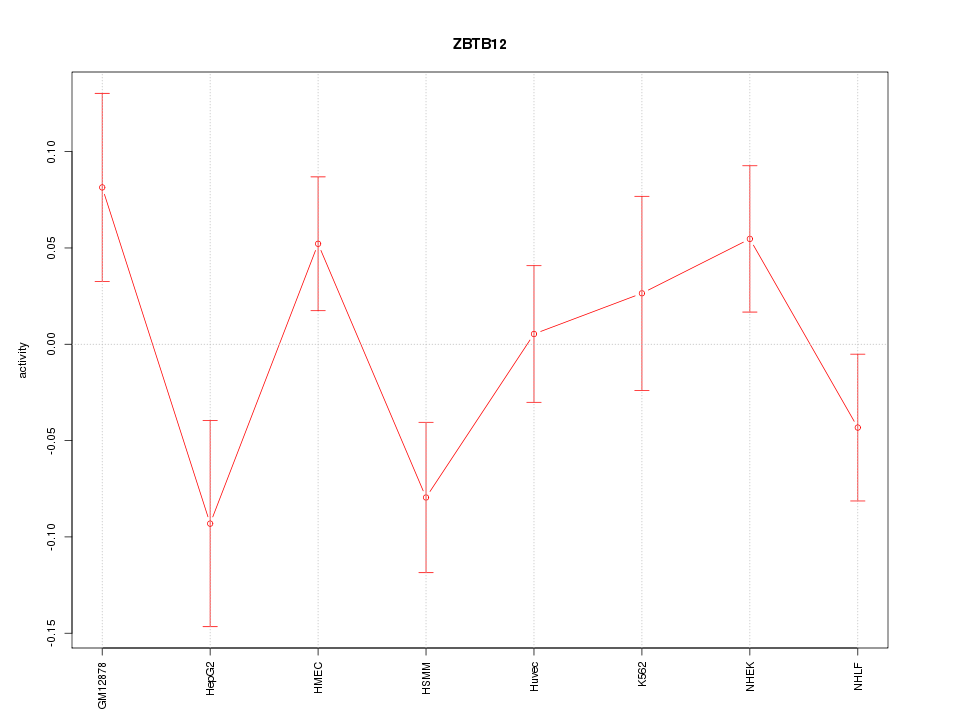
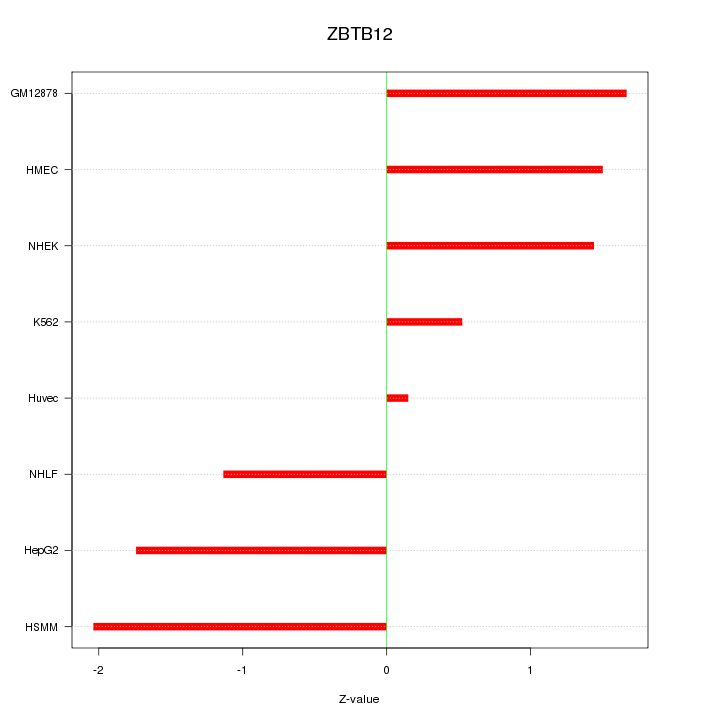
Motif ID: ZBTB12
Z-value: 1.409
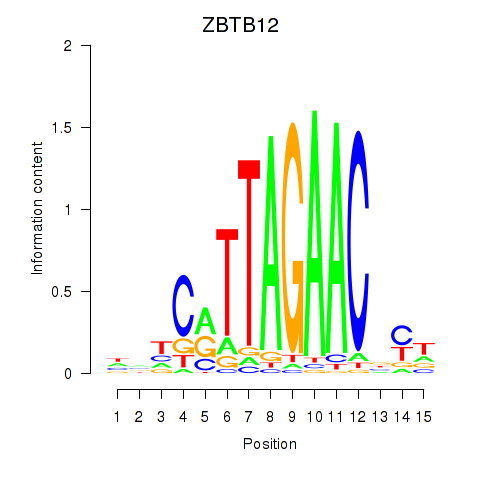
Transcription factors associated with ZBTB12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZBTB12 | ENSG00000204366.3 | ZBTB12 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.3 | 1.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.6 | GO:0061198 | fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.2 | 4.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 0.6 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.4 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.1 | 2.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.6 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.2 | GO:2000078 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 1.8 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 2.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.1 | GO:0000236 | mitotic prometaphase(GO:0000236) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 3.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.6 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 2.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.2 | GO:0008199 | acid phosphatase activity(GO:0003993) ferric iron binding(GO:0008199) |
| 0.1 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 1.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.0 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0016594 | glycine binding(GO:0016594) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |