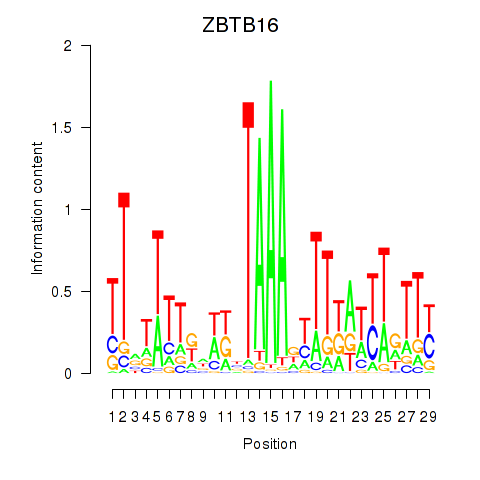
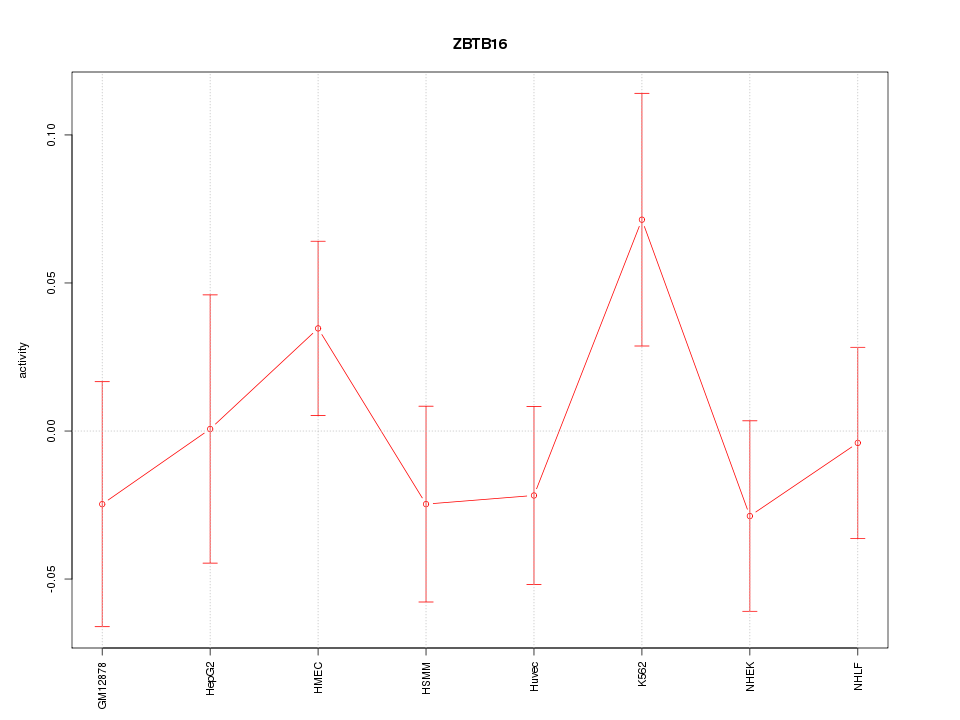
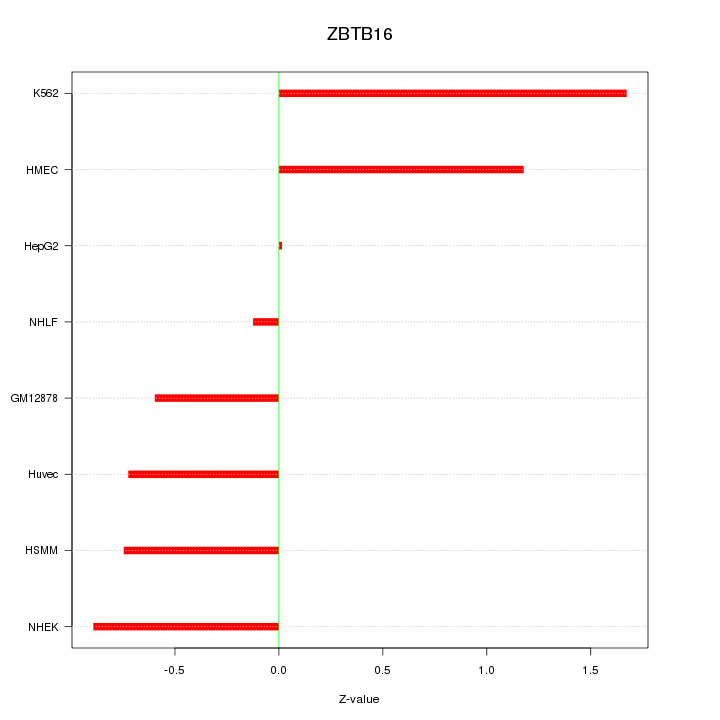
| 0.3 |
2.0 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 |
0.7 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 |
0.5 |
GO:0021570 |
rhombomere 4 development(GO:0021570) |
| 0.2 |
0.9 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
0.4 |
GO:0070340 |
cell surface pattern recognition receptor signaling pathway(GO:0002752) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 |
0.3 |
GO:0071460 |
negative regulation of necrotic cell death(GO:0060547) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 |
1.4 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 |
0.3 |
GO:0071025 |
intracellular mRNA localization(GO:0008298) RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 |
0.2 |
GO:0019441 |
tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 |
0.4 |
GO:0042448 |
progesterone metabolic process(GO:0042448) |
| 0.0 |
0.3 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.0 |
0.3 |
GO:0019264 |
L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 |
0.7 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.0 |
0.6 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.4 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
1.2 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 |
0.6 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.7 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.3 |
GO:0015810 |
C4-dicarboxylate transport(GO:0015740) aspartate transport(GO:0015810) |
| 0.0 |
0.1 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.0 |
0.1 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.2 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
0.6 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.0 |
0.4 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.4 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.7 |
GO:0034968 |
histone lysine methylation(GO:0034968) |
| 0.0 |
0.7 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.3 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.3 |
GO:0075713 |
establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 |
0.0 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.0 |
0.2 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
0.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.0 |
GO:0002887 |
negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 |
0.2 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 |
0.5 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 |
0.3 |
GO:0006693 |
prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.1 |
GO:0009750 |
response to fructose(GO:0009750) |
| 0.0 |
0.0 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |