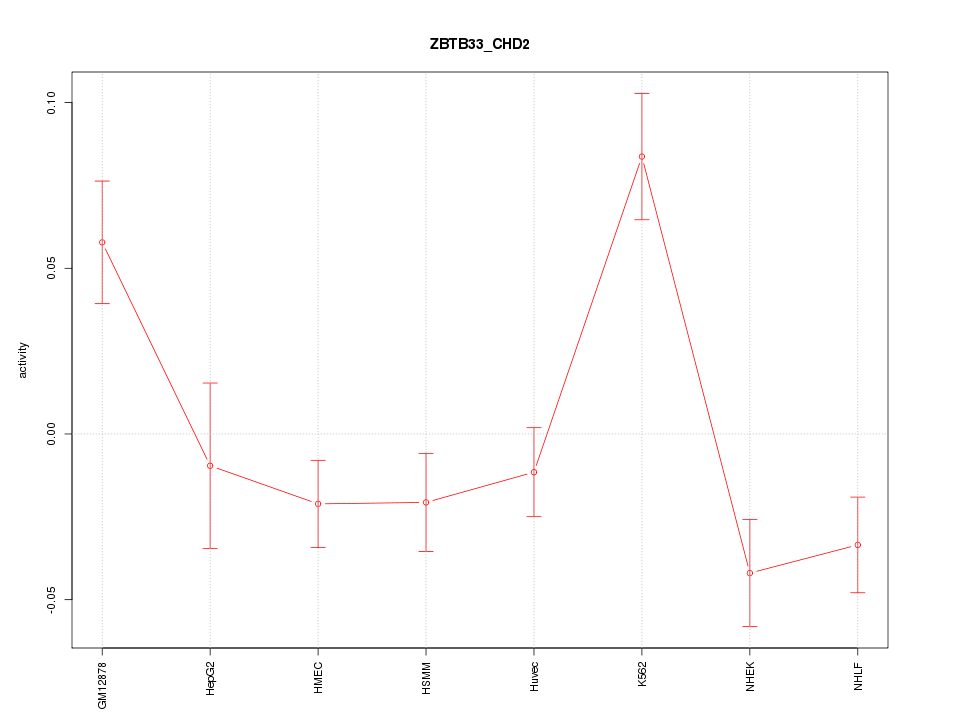
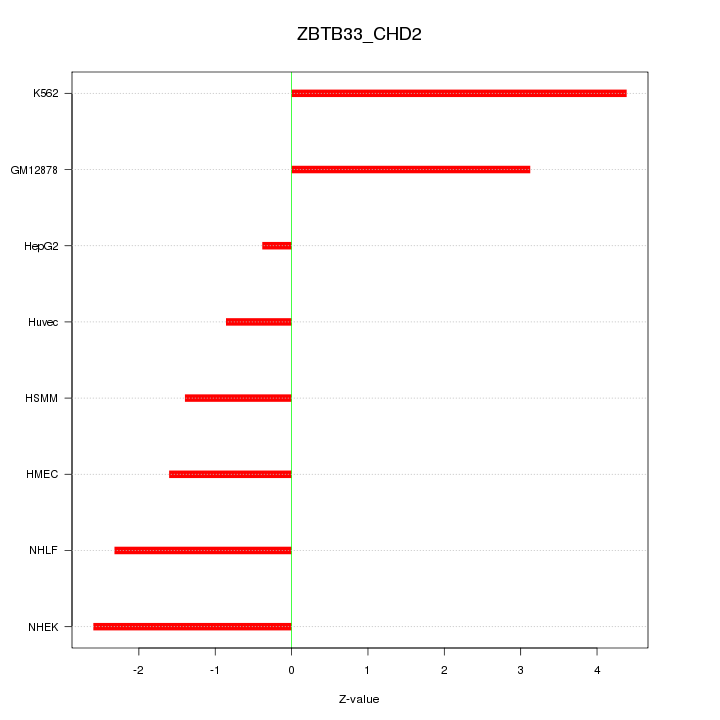
Motif ID: ZBTB33_CHD2
Z-value: 2.412
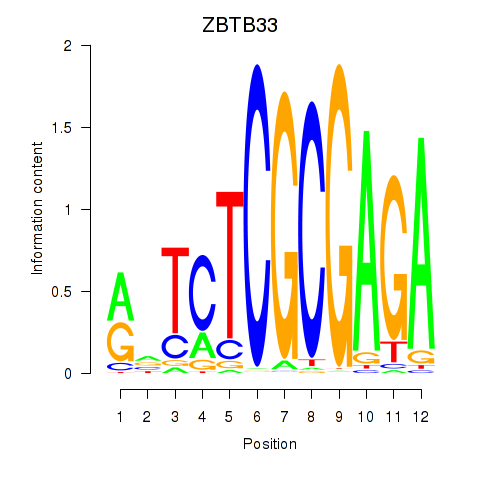
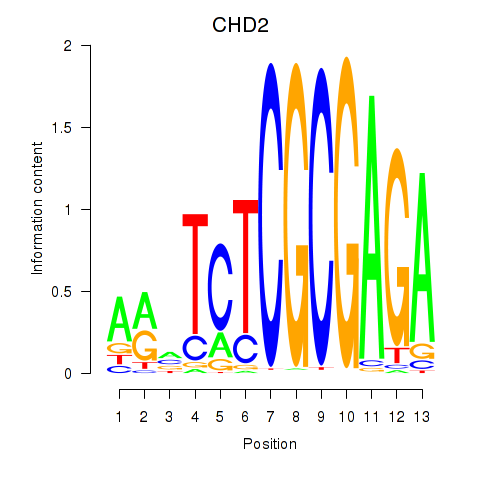
Transcription factors associated with ZBTB33_CHD2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CHD2 | ENSG00000173575.14 | CHD2 |
| ZBTB33 | ENSG00000177485.6 | ZBTB33 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.8 | 0.8 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.6 | 0.6 | GO:0033002 | muscle cell proliferation(GO:0033002) smooth muscle cell proliferation(GO:0048659) |
| 0.6 | 3.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.6 | 3.5 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.5 | 1.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.5 | 2.7 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.4 | 2.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.4 | 1.2 | GO:0060547 | negative regulation of necrotic cell death(GO:0060547) |
| 0.3 | 6.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.3 | 3.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 4.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 1.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.3 | 1.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.3 | 1.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) cleavage involved in rRNA processing(GO:0000469) endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) RNA 5'-end processing(GO:0000966) RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.3 | 1.0 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.3 | 0.8 | GO:0043418 | cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) |
| 0.2 | 1.5 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.2 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 0.9 | GO:0035283 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 2.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.2 | 0.7 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 0.7 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.2 | 2.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.1 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.2 | 1.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 1.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.2 | 0.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 | 1.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.2 | 1.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.2 | 0.5 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) nuclear-transcribed mRNA catabolic process(GO:0000956) mRNA catabolic process(GO:0006402) |
| 0.2 | 0.5 | GO:1990170 | detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.2 | 4.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.5 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.2 | 0.9 | GO:0015853 | adenine transport(GO:0015853) |
| 0.2 | 2.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.4 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.4 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.5 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.4 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.7 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 2.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.7 | GO:0060412 | ventricular septum morphogenesis(GO:0060412) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.3 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.5 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 2.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.1 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.1 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.1 | 0.4 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.9 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.5 | GO:0033504 | floor plate development(GO:0033504) |
| 0.1 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.5 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.9 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.4 | GO:0016265 | obsolete death(GO:0016265) |
| 0.1 | 0.3 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.5 | GO:0001823 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 0.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.1 | GO:0090042 | tubulin deacetylation(GO:0090042) |
| 0.1 | 0.5 | GO:1900087 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 | 1.1 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 3.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 4.4 | GO:0000236 | mitotic prometaphase(GO:0000236) |
| 0.0 | 0.3 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.7 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0046668 | retinal cell programmed cell death(GO:0046666) regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 1.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.8 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.2 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.2 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.5 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.0 | 0.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.9 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.3 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 1.2 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.5 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.1 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 3.0 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.4 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.3 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.3 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 1.0 | GO:0043038 | tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.2 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.0 | 0.3 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 3.2 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.0 | 0.8 | GO:0048593 | camera-type eye morphogenesis(GO:0048593) |
| 0.0 | 0.3 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.3 | GO:0016070 | RNA metabolic process(GO:0016070) |
| 0.0 | 0.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.9 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 1.9 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.4 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 2.8 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:0051276 | chromosome organization(GO:0051276) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.3 | GO:0009416 | response to light stimulus(GO:0009416) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0046070 | dGTP metabolic process(GO:0046070) purine deoxyribonucleoside metabolic process(GO:0046122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0000802 | transverse filament(GO:0000802) |
| 0.6 | 6.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.5 | 4.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.4 | 3.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 0.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.3 | 1.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 3.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 2.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) INO80-type complex(GO:0097346) |
| 0.1 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.5 | GO:0019815 | immunoglobulin complex(GO:0019814) B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.7 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.8 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.1 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 2.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.7 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.9 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 1.1 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 11.5 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 3.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0031968 | organelle outer membrane(GO:0031968) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.5 | GO:1990904 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.6 | 2.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.6 | 2.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.6 | 3.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 1.6 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.5 | 2.6 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 1.3 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.4 | 1.2 | GO:0015217 | purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 3.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 1.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.3 | 6.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 0.8 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 0.7 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.9 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.9 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.2 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.5 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 0.5 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.2 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 3.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 0.7 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.5 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 1.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 2.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 1.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.5 | GO:0017163 | obsolete basal transcription repressor activity(GO:0017163) |
| 0.1 | 0.3 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.1 | 0.6 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 3.8 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.5 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.3 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0017096 | acetylserotonin O-methyltransferase activity(GO:0017096) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 1.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.1 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.9 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |