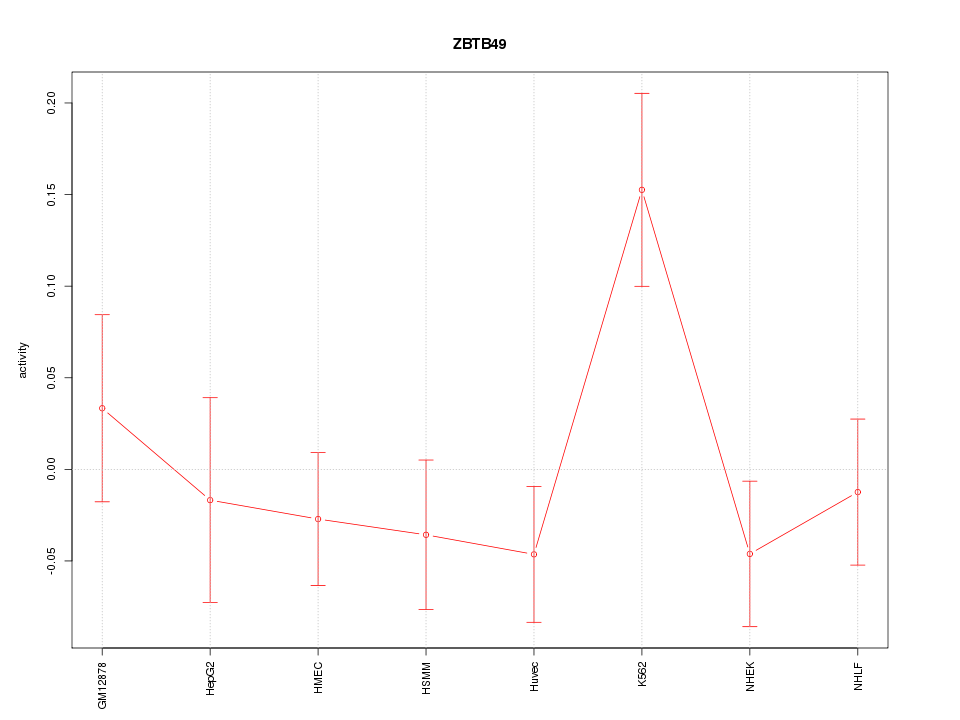
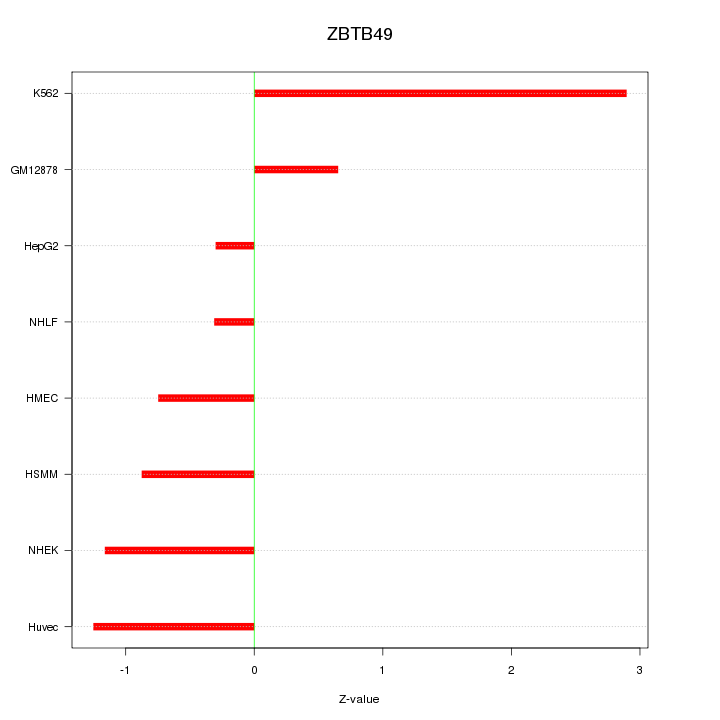
Motif ID: ZBTB49
Z-value: 1.287
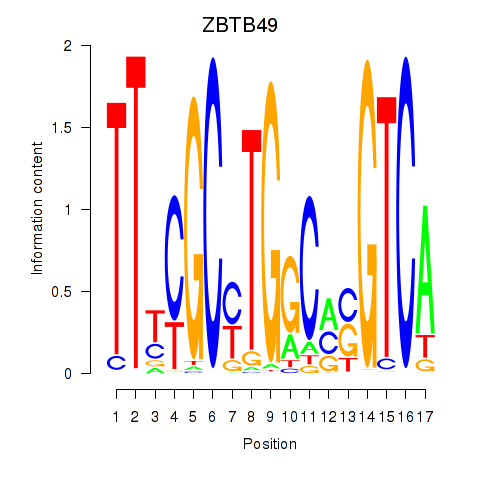
Transcription factors associated with ZBTB49:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZBTB49 | ENSG00000168826.11 | ZBTB49 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.7 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.3 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.6 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.1 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.1 | 1.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0034661 | ncRNA catabolic process(GO:0034661) |
| 0.0 | 1.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.6 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 2.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.0 | GO:1990204 | oxidoreductase complex(GO:1990204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.3 | GO:0004519 | endonuclease activity(GO:0004519) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |