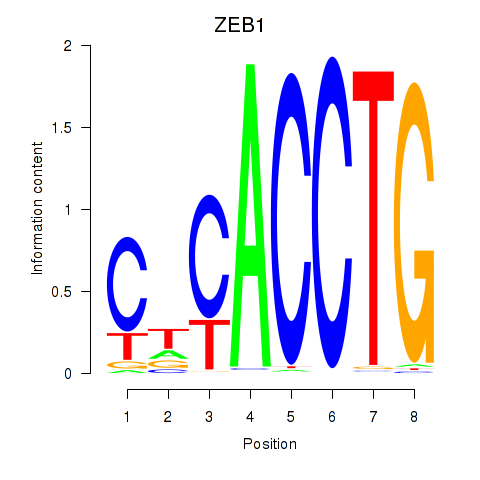
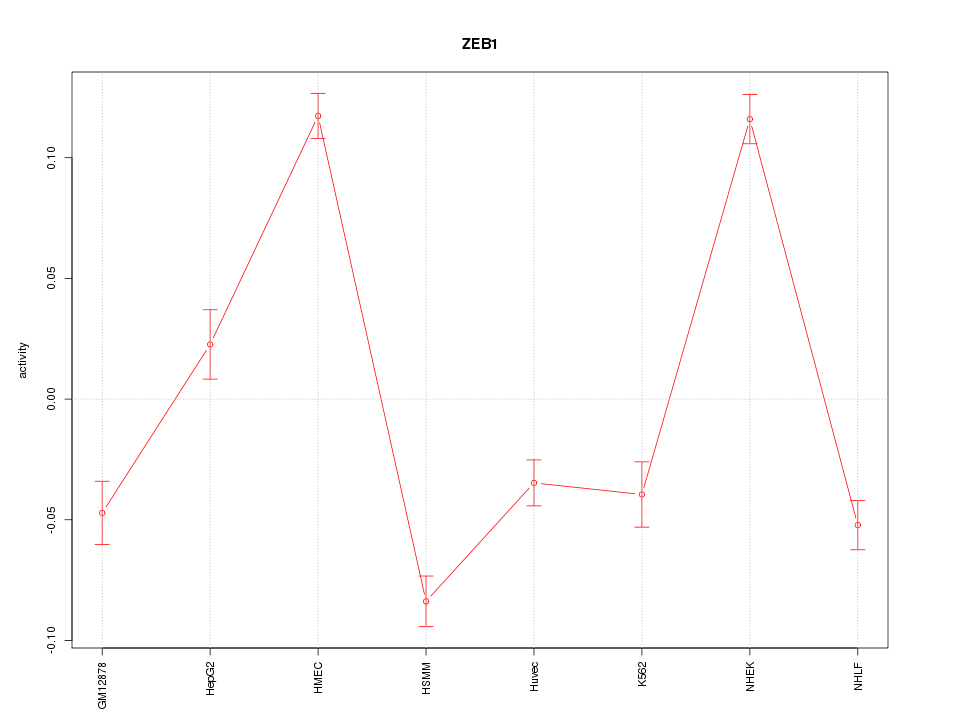
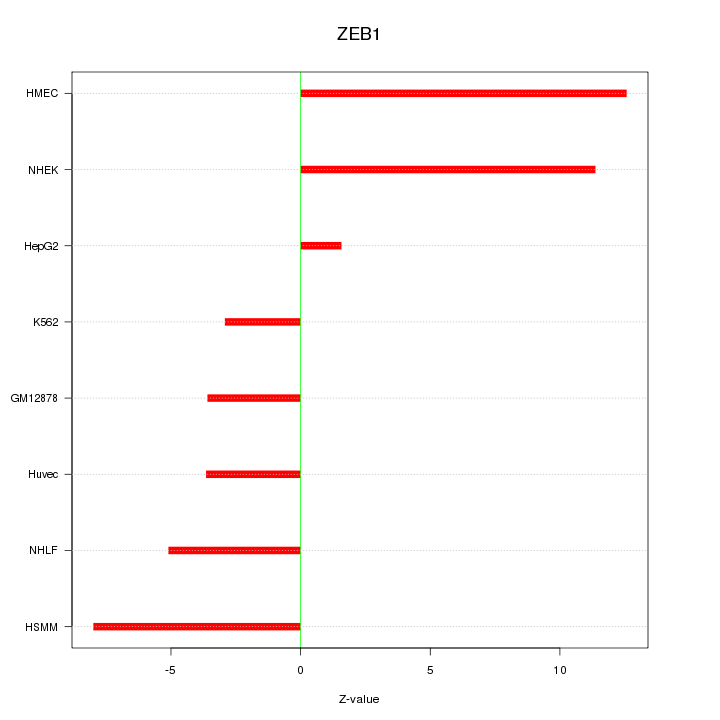
| 5.1 |
15.3 |
GO:0003308 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 3.7 |
11.0 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 3.6 |
10.9 |
GO:0018243 |
protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 3.6 |
10.9 |
GO:0048627 |
myoblast development(GO:0048627) |
| 3.5 |
14.0 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 3.2 |
22.2 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 2.9 |
5.8 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 2.6 |
2.6 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 2.2 |
64.4 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 2.1 |
6.2 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 2.0 |
8.0 |
GO:0042231 |
interleukin-13 biosynthetic process(GO:0042231) |
| 1.7 |
5.2 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 1.7 |
5.2 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.7 |
6.6 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.4 |
5.6 |
GO:0070933 |
histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 1.3 |
22.9 |
GO:0043616 |
keratinocyte proliferation(GO:0043616) |
| 1.3 |
5.2 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 1.2 |
6.1 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 1.2 |
3.6 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 1.2 |
5.9 |
GO:0044854 |
membrane raft assembly(GO:0001765) nitric oxide homeostasis(GO:0033484) negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 1.2 |
3.5 |
GO:0009635 |
response to herbicide(GO:0009635) |
| 1.1 |
2.2 |
GO:0031394 |
regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.1 |
9.9 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 1.1 |
3.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.1 |
5.4 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 1.0 |
3.1 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.0 |
1.0 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 1.0 |
10.7 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 1.0 |
3.9 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.9 |
3.7 |
GO:0032849 |
positive regulation of cellular pH reduction(GO:0032849) |
| 0.9 |
4.5 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.9 |
2.7 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 |
1.7 |
GO:0051547 |
regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.9 |
9.5 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.9 |
10.3 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.8 |
44.7 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.8 |
8.4 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.8 |
3.3 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.8 |
4.0 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.8 |
23.5 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.7 |
4.5 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.7 |
21.7 |
GO:0030216 |
keratinocyte differentiation(GO:0030216) |
| 0.7 |
7.9 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.7 |
2.0 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.7 |
2.0 |
GO:0046368 |
GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.6 |
2.6 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.6 |
1.3 |
GO:0060061 |
Spemann organizer formation(GO:0060061) |
| 0.6 |
0.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
1.9 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.6 |
3.0 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.6 |
1.8 |
GO:0070092 |
regulation of glucagon secretion(GO:0070092) |
| 0.6 |
3.5 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 |
0.5 |
GO:0045844 |
positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.5 |
1.6 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 |
0.5 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.5 |
1.5 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.5 |
1.0 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.5 |
11.4 |
GO:0060444 |
branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.5 |
3.5 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.5 |
2.0 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) regulation of phosphatidylinositol biosynthetic process(GO:0010511) obsolete regulation of calcium ion transport via store-operated calcium channel activity(GO:0032234) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.5 |
1.0 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.5 |
0.9 |
GO:0032226 |
positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.5 |
1.4 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.5 |
4.1 |
GO:1902743 |
regulation of lamellipodium organization(GO:1902743) |
| 0.5 |
1.4 |
GO:0045978 |
negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.4 |
0.4 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.4 |
0.9 |
GO:0072201 |
negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.4 |
2.2 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.4 |
11.9 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.4 |
2.5 |
GO:0034201 |
response to oleic acid(GO:0034201) |
| 0.4 |
2.5 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.4 |
2.1 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.4 |
30.1 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.4 |
52.0 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 |
9.6 |
GO:0060512 |
prostate gland morphogenesis(GO:0060512) |
| 0.4 |
2.4 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.4 |
1.2 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.4 |
0.4 |
GO:0033088 |
negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 |
3.5 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.4 |
2.7 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.4 |
30.5 |
GO:0001889 |
liver development(GO:0001889) |
| 0.4 |
5.4 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.4 |
1.9 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.4 |
1.5 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.4 |
0.7 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 |
17.1 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.4 |
10.0 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.4 |
2.8 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.4 |
6.0 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 |
1.1 |
GO:0034635 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 |
3.5 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.3 |
2.4 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 |
1.3 |
GO:0072180 |
nephric duct development(GO:0072176) mesonephric duct development(GO:0072177) nephric duct morphogenesis(GO:0072178) mesonephric duct morphogenesis(GO:0072180) |
| 0.3 |
1.7 |
GO:0042159 |
lipoprotein catabolic process(GO:0042159) |
| 0.3 |
1.0 |
GO:0071476 |
response to vitamin B1(GO:0010266) cellular hypotonic response(GO:0071476) |
| 0.3 |
1.0 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.3 |
4.5 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.3 |
1.3 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.3 |
0.9 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.3 |
2.5 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.3 |
0.9 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 |
1.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 |
0.3 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.3 |
0.9 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 0.3 |
0.9 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 |
1.1 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.3 |
8.9 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.3 |
5.3 |
GO:0050927 |
positive regulation of positive chemotaxis(GO:0050927) |
| 0.3 |
0.6 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.3 |
0.8 |
GO:0048878 |
chemical homeostasis(GO:0048878) |
| 0.3 |
0.3 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.3 |
1.6 |
GO:0046146 |
tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 |
1.0 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 |
2.0 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.2 |
30.9 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 |
1.5 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.2 |
0.5 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.2 |
0.2 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 |
0.2 |
GO:0048659 |
smooth muscle cell proliferation(GO:0048659) |
| 0.2 |
1.4 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 |
1.4 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.2 |
1.1 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 |
1.1 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.2 |
0.7 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.2 |
1.8 |
GO:0051005 |
negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 |
0.7 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 |
0.2 |
GO:0051919 |
positive regulation of fibrinolysis(GO:0051919) |
| 0.2 |
0.6 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.2 |
3.0 |
GO:0035315 |
hair cell differentiation(GO:0035315) |
| 0.2 |
1.3 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
0.2 |
GO:0090205 |
positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.2 |
1.9 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 |
0.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.2 |
0.4 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.2 |
0.6 |
GO:0010041 |
response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.2 |
0.4 |
GO:0055078 |
sodium ion homeostasis(GO:0055078) |
| 0.2 |
2.6 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.2 |
0.8 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 |
0.2 |
GO:0072053 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.2 |
1.4 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 |
0.2 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 |
0.6 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.2 |
2.1 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 |
0.8 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 |
0.6 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 |
1.8 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.2 |
2.2 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 |
4.8 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 |
1.2 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 |
1.1 |
GO:0006935 |
chemotaxis(GO:0006935) taxis(GO:0042330) |
| 0.2 |
2.3 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.2 |
1.0 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 |
1.6 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.2 |
0.5 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.2 |
2.4 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 |
0.8 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.2 |
1.2 |
GO:0007549 |
dosage compensation(GO:0007549) |
| 0.2 |
0.5 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.2 |
5.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.2 |
0.8 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.2 |
0.6 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 |
0.3 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 |
0.5 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
1.2 |
GO:0048010 |
vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 |
0.5 |
GO:0050685 |
positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.2 |
1.2 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.2 |
0.9 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 |
0.5 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting two-sector ATPase complex assembly(GO:0070071) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
3.6 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 |
0.4 |
GO:0000050 |
urea cycle(GO:0000050) |
| 0.1 |
1.6 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 |
0.3 |
GO:0042311 |
vasodilation(GO:0042311) |
| 0.1 |
0.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.1 |
0.6 |
GO:0010466 |
negative regulation of peptidase activity(GO:0010466) |
| 0.1 |
0.4 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 |
1.4 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
0.3 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.1 |
GO:0044764 |
multi-organism cellular process(GO:0044764) |
| 0.1 |
2.9 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.6 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 |
0.8 |
GO:0042362 |
fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 |
0.3 |
GO:0003077 |
obsolete negative regulation of diuresis(GO:0003077) |
| 0.1 |
1.8 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.1 |
0.4 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 |
3.2 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.1 |
1.2 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
1.1 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.1 |
0.7 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.5 |
GO:0019673 |
GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 |
2.0 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.1 |
0.4 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
4.3 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 |
1.0 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 |
1.0 |
GO:0032375 |
negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.1 |
0.9 |
GO:0016559 |
peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.1 |
2.1 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.1 |
0.4 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 |
1.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
0.4 |
GO:0048075 |
regulation of eye pigmentation(GO:0048073) positive regulation of eye pigmentation(GO:0048075) |
| 0.1 |
0.5 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) positive regulation of epidermis development(GO:0045684) positive regulation of hair follicle development(GO:0051798) |
| 0.1 |
1.0 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.1 |
0.2 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 |
2.0 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.1 |
0.4 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 |
0.3 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.1 |
0.3 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.1 |
2.1 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.4 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 |
0.2 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
1.2 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.3 |
GO:0006356 |
regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 |
0.3 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.1 |
0.7 |
GO:1902667 |
regulation of axon extension involved in axon guidance(GO:0048841) regulation of axon guidance(GO:1902667) |
| 0.1 |
6.6 |
GO:0007163 |
establishment or maintenance of cell polarity(GO:0007163) |
| 0.1 |
0.4 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) regulation of cardiocyte differentiation(GO:1905207) |
| 0.1 |
0.4 |
GO:0032070 |
regulation of deoxyribonuclease activity(GO:0032070) |
| 0.1 |
0.3 |
GO:0042727 |
flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 |
0.5 |
GO:0030858 |
positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.1 |
0.7 |
GO:0061037 |
negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) |
| 0.1 |
0.2 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 |
0.3 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.1 |
0.3 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 |
0.6 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
0.5 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.1 |
1.0 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 |
0.9 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.1 |
1.1 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 |
0.6 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 |
0.3 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 |
0.3 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
1.4 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.1 |
0.6 |
GO:0032786 |
positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 |
0.3 |
GO:0060295 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium-dependent cell motility(GO:0060285) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of microtubule-based movement(GO:0060632) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 |
1.3 |
GO:0007249 |
I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 |
0.3 |
GO:0070633 |
transepithelial transport(GO:0070633) |
| 0.1 |
1.6 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 |
4.1 |
GO:0051084 |
'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 |
0.8 |
GO:0055075 |
potassium ion homeostasis(GO:0055075) |
| 0.1 |
0.3 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.7 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
1.9 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.6 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 |
0.2 |
GO:0048875 |
surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.1 |
0.5 |
GO:0055093 |
response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.1 |
0.3 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.1 |
0.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.1 |
0.2 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.1 |
0.2 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.1 |
0.4 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
0.3 |
GO:0060363 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cranial suture morphogenesis(GO:0060363) cellular response to sterol depletion(GO:0071501) craniofacial suture morphogenesis(GO:0097094) |
| 0.1 |
0.8 |
GO:0006702 |
androgen biosynthetic process(GO:0006702) |
| 0.1 |
1.0 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 |
0.9 |
GO:0050704 |
regulation of interleukin-1 secretion(GO:0050704) positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 |
1.6 |
GO:0016042 |
lipid catabolic process(GO:0016042) |
| 0.1 |
0.2 |
GO:0032621 |
interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 |
0.2 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 |
3.2 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.1 |
0.4 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.1 |
1.6 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.4 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.1 |
0.2 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.1 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.1 |
0.3 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
2.0 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 |
0.3 |
GO:0051852 |
disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.1 |
1.1 |
GO:0006954 |
inflammatory response(GO:0006954) |
| 0.1 |
0.7 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.3 |
GO:0043030 |
regulation of macrophage activation(GO:0043030) |
| 0.1 |
2.8 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
3.1 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.1 |
0.2 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.1 |
1.0 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 |
0.6 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 |
0.1 |
GO:0048715 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.1 |
0.2 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.1 |
0.2 |
GO:0051608 |
histamine transport(GO:0051608) |
| 0.1 |
0.1 |
GO:0032958 |
inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 |
0.1 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 |
0.4 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.1 |
0.2 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 |
0.2 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.1 |
1.8 |
GO:0007613 |
memory(GO:0007613) |
| 0.1 |
1.7 |
GO:0040014 |
regulation of multicellular organism growth(GO:0040014) |
| 0.1 |
0.2 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.1 |
0.2 |
GO:0060123 |
regulation of growth hormone secretion(GO:0060123) positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 |
0.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.2 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.4 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 |
1.0 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 |
0.3 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
1.7 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.1 |
0.2 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 |
1.7 |
GO:0006672 |
ceramide metabolic process(GO:0006672) |
| 0.1 |
0.4 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.1 |
1.2 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.1 |
0.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.4 |
GO:0002860 |
natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 |
0.9 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.7 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0051216 |
cartilage development(GO:0051216) |
| 0.0 |
0.4 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.0 |
0.7 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.0 |
0.2 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 |
0.4 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.4 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
0.2 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.1 |
GO:0034374 |
low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 |
0.9 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |
| 0.0 |
0.3 |
GO:0035116 |
embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 |
0.5 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.2 |
GO:0045920 |
negative regulation of exocytosis(GO:0045920) |
| 0.0 |
0.6 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.1 |
GO:0016445 |
somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 |
0.3 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
0.0 |
GO:0090594 |
wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 |
2.2 |
GO:0042552 |
myelination(GO:0042552) |
| 0.0 |
3.3 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.6 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.3 |
GO:0071482 |
cellular response to light stimulus(GO:0071482) |
| 0.0 |
3.8 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.2 |
GO:0021615 |
glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
5.2 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.1 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.2 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.7 |
GO:0016101 |
retinoid metabolic process(GO:0001523) diterpenoid metabolic process(GO:0016101) |
| 0.0 |
2.3 |
GO:0008544 |
epidermis development(GO:0008544) |
| 0.0 |
0.1 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.0 |
0.1 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.9 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.9 |
GO:0006909 |
phagocytosis(GO:0006909) |
| 0.0 |
0.2 |
GO:0015851 |
nucleobase transport(GO:0015851) |
| 0.0 |
0.1 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.1 |
GO:0006200 |
obsolete ATP catabolic process(GO:0006200) |
| 0.0 |
0.3 |
GO:0015780 |
nucleotide-sugar transport(GO:0015780) |
| 0.0 |
0.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.1 |
GO:0015893 |
drug transport(GO:0015893) |
| 0.0 |
0.7 |
GO:0050819 |
negative regulation of coagulation(GO:0050819) |
| 0.0 |
0.1 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
1.7 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
1.0 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.2 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
1.5 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.3 |
GO:0006928 |
movement of cell or subcellular component(GO:0006928) |
| 0.0 |
0.1 |
GO:0021988 |
olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 |
2.4 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.2 |
GO:0050806 |
positive regulation of synaptic transmission(GO:0050806) |
| 0.0 |
0.1 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.0 |
0.2 |
GO:1901998 |
tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0018283 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 |
0.4 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.0 |
1.7 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
1.6 |
GO:0006898 |
receptor-mediated endocytosis(GO:0006898) |
| 0.0 |
0.3 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.5 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.0 |
0.2 |
GO:0043954 |
cell junction maintenance(GO:0034331) cellular component maintenance(GO:0043954) cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.5 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.6 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.8 |
GO:0006754 |
ATP biosynthetic process(GO:0006754) |
| 0.0 |
0.0 |
GO:0015802 |
basic amino acid transport(GO:0015802) |
| 0.0 |
0.1 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.0 |
0.1 |
GO:0031657 |
regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 |
0.0 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.0 |
0.2 |
GO:0006555 |
methionine metabolic process(GO:0006555) |
| 0.0 |
0.0 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
0.5 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.9 |
GO:0007338 |
single fertilization(GO:0007338) |
| 0.0 |
0.4 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.0 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.3 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.3 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.1 |
GO:0002026 |
regulation of the force of heart contraction(GO:0002026) |
| 0.0 |
0.1 |
GO:0046210 |
nitric oxide catabolic process(GO:0046210) |
| 0.0 |
0.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.4 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.3 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.2 |
GO:0015838 |
quaternary ammonium group transport(GO:0015697) amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 |
0.3 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0030878 |
thyroid gland development(GO:0030878) |
| 0.0 |
0.0 |
GO:0032025 |
response to cobalt ion(GO:0032025) |
| 0.0 |
0.1 |
GO:0030826 |
regulation of cGMP biosynthetic process(GO:0030826) |
| 0.0 |
0.4 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.8 |
GO:0043542 |
endothelial cell migration(GO:0043542) |
| 0.0 |
0.4 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.1 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.2 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.4 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.3 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.2 |
GO:1901019 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) regulation of calcium ion transmembrane transporter activity(GO:1901019) regulation of cation channel activity(GO:2001257) |
| 0.0 |
0.2 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.1 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
0.0 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0001947 |
heart looping(GO:0001947) embryonic heart tube morphogenesis(GO:0003143) determination of heart left/right asymmetry(GO:0061371) |
| 0.0 |
0.1 |
GO:0048665 |
neuron fate specification(GO:0048665) |
| 0.0 |
0.0 |
GO:0072202 |
cell differentiation involved in metanephros development(GO:0072202) |
| 0.0 |
0.3 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.1 |
GO:0016075 |
rRNA catabolic process(GO:0016075) |
| 0.0 |
0.3 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.5 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) ERBB signaling pathway(GO:0038127) |
| 0.0 |
0.3 |
GO:0009880 |
embryonic pattern specification(GO:0009880) |
| 0.0 |
0.2 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.1 |
GO:0046546 |
male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 |
0.2 |
GO:0042417 |
dopamine metabolic process(GO:0042417) |
| 0.0 |
0.0 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.1 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 |
0.1 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.0 |
0.2 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.3 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.1 |
GO:0045926 |
negative regulation of growth(GO:0045926) |
| 0.0 |
0.1 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.0 |
0.1 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 |
0.2 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.1 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:0008306 |
associative learning(GO:0008306) |
| 0.0 |
0.6 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |