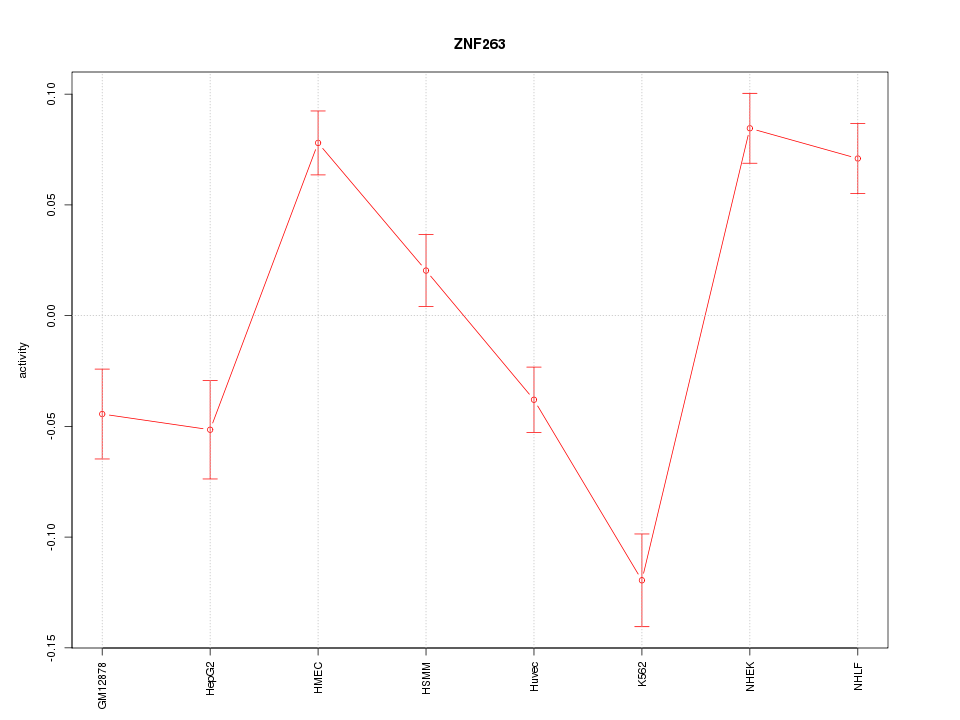
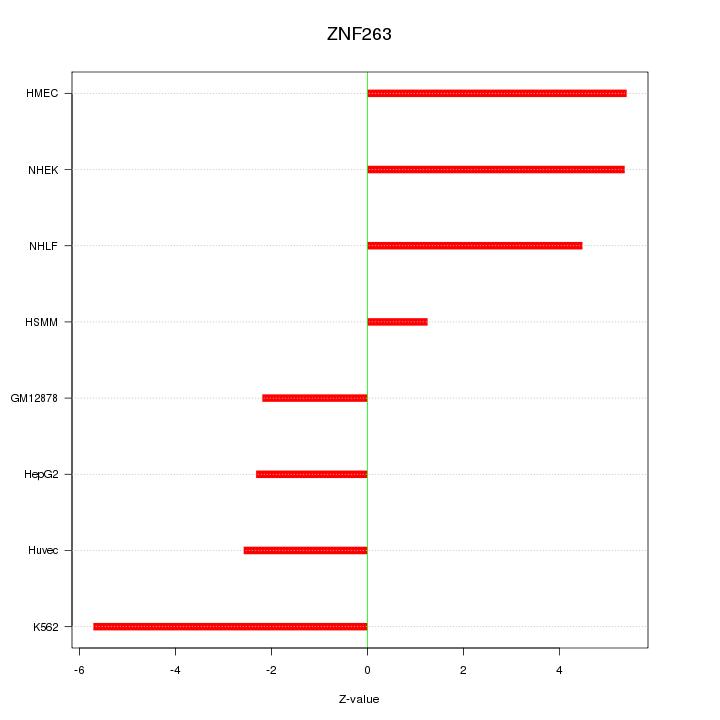
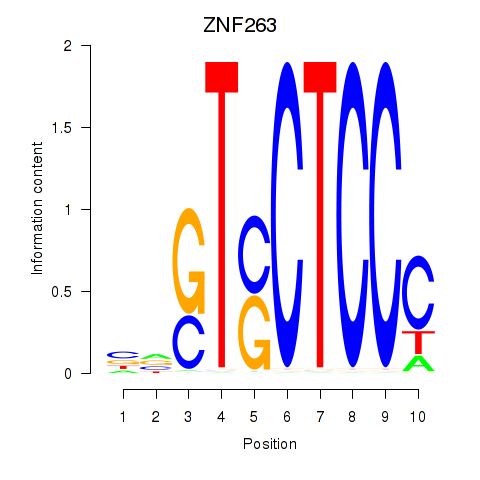
Motif ID: ZNF263
Z-value: 4.015
Transcription factors associated with ZNF263:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZNF263 | ENSG00000006194.6 | ZNF263 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 2.3 | 13.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 2.3 | 13.8 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 2.0 | 14.0 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 1.9 | 13.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.2 | 3.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 1.2 | 6.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 1.1 | 4.4 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 1.1 | 3.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.0 | 3.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.0 | 6.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.9 | 2.7 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 0.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.8 | 6.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.8 | 5.4 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.7 | 1.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.7 | 2.9 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.7 | 2.1 | GO:0035987 | endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.7 | 2.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.7 | 8.0 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.6 | 5.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.6 | 2.4 | GO:2001258 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of cation channel activity(GO:2001258) |
| 0.6 | 1.8 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.6 | 1.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.6 | 1.7 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.6 | 1.7 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.5 | 1.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.5 | 2.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 3.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 3.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.5 | 1.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.5 | 1.4 | GO:0031133 | regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.4 | 12.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 4.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 9.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 1.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 1.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 1.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.3 | 2.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.3 | 1.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.3 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 0.9 | GO:0021834 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.3 | 0.9 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.8 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 | 1.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.3 | 6.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.3 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.3 | 1.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 2.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.3 | 0.8 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.3 | 0.8 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.3 | 1.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) proline transport(GO:0015824) |
| 0.2 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 8.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 0.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 0.7 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.2 | 1.6 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 3.6 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.2 | 5.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 0.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 1.3 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.2 | 0.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 0.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 0.6 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.2 | 0.6 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.2 | 0.7 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.2 | 2.2 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.2 | 0.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 2.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 1.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 4.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 2.6 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.2 | 1.1 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.2 | 0.3 | GO:0060024 | musculoskeletal movement, spinal reflex action(GO:0050883) rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 1.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.2 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 4.7 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.2 | 2.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 0.5 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.6 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.4 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 7.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.7 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.1 | 0.7 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 1.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 1.0 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 2.2 | GO:0016049 | cell growth(GO:0016049) |
| 0.1 | 0.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 7.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 7.2 | GO:0008064 | regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 | 2.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 2.8 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.1 | 0.6 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.2 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.1 | 1.5 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 1.7 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 2.6 | GO:0006029 | proteoglycan metabolic process(GO:0006029) |
| 0.1 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.3 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.1 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.4 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 13.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.1 | 1.0 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 6.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.2 | GO:0048668 | collateral sprouting(GO:0048668) regulation of collateral sprouting(GO:0048670) |
| 0.1 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.4 | GO:0009629 | response to gravity(GO:0009629) |
| 0.1 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:0072369 | low-density lipoprotein particle receptor biosynthetic process(GO:0045713) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 | 5.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.2 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.1 | 0.1 | GO:0014048 | regulation of glutamate secretion(GO:0014048) |
| 0.1 | 0.8 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.1 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.1 | 5.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 1.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 3.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 3.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) mesenchymal to epithelial transition(GO:0060231) renal vesicle morphogenesis(GO:0072077) renal vesicle development(GO:0072087) metanephric nephron morphogenesis(GO:0072273) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 1.2 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.7 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.3 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.6 | GO:0001975 | response to amphetamine(GO:0001975) |
| 0.0 | 0.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.6 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.0 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.0 | 4.9 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.8 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.1 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 4.4 | GO:0051325 | interphase(GO:0051325) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 1.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.3 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0061339 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) cellular biogenic amine biosynthetic process(GO:0042401) |
| 0.0 | 0.4 | GO:0035050 | embryonic heart tube development(GO:0035050) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.4 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.9 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 2.8 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.7 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.4 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.4 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0071772 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.0 | GO:0007285 | primary spermatocyte growth(GO:0007285) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0045844 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 0.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.0 | 8.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.8 | 6.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.6 | 6.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 9.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.6 | 6.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.5 | 3.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.5 | 7.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.5 | 4.7 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 1.8 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.4 | 2.3 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.3 | 2.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 4.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 1.6 | GO:0032449 | CBM complex(GO:0032449) |
| 0.3 | 5.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 5.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.2 | GO:0019718 | obsolete rough microsome(GO:0019718) |
| 0.2 | 2.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 3.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 0.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.2 | 1.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 1.1 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.2 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 15.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 10.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 4.9 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 4.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 7.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.9 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0060170 | BBSome(GO:0034464) ciliary membrane(GO:0060170) |
| 0.0 | 8.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 20.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.7 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0043292 | myofibril(GO:0030016) contractile fiber(GO:0043292) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 15.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.9 | 13.3 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 1.1 | 13.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 1.0 | 3.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.0 | 6.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 2.9 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.9 | 2.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.9 | 8.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.9 | 1.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.9 | 2.6 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.9 | 8.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.8 | 2.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 4.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.6 | 1.7 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.6 | 2.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.5 | 1.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 2.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 1.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.5 | 6.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 7.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 2.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 1.2 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.4 | 15.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 1.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.4 | 3.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.4 | 4.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 1.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 1.4 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.3 | 2.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 1.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.3 | 3.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 0.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 8.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 1.8 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 1.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.3 | 1.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 4.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.4 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.2 | 1.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.2 | 5.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 3.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 3.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 5.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 0.9 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.2 | 4.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 11.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 0.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.0 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.2 | 1.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 2.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.2 | 1.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 1.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 2.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 2.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.0 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.1 | 4.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.0 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 11.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.8 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 1.6 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.5 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 9.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 3.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.9 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 1.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.3 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 6.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 4.7 | GO:0042379 | chemokine receptor binding(GO:0042379) |
| 0.1 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 5.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 14.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 3.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 7.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 3.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) BH domain binding(GO:0051400) |
| 0.1 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 26.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.5 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 1.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 4.1 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 3.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 13.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0001104 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 3.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 1.6 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.7 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 11.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 6.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 9.3 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 0.7 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.7 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.2 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 0.7 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.1 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.3 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | ST_ADRENERGIC | Adrenergic Pathway |