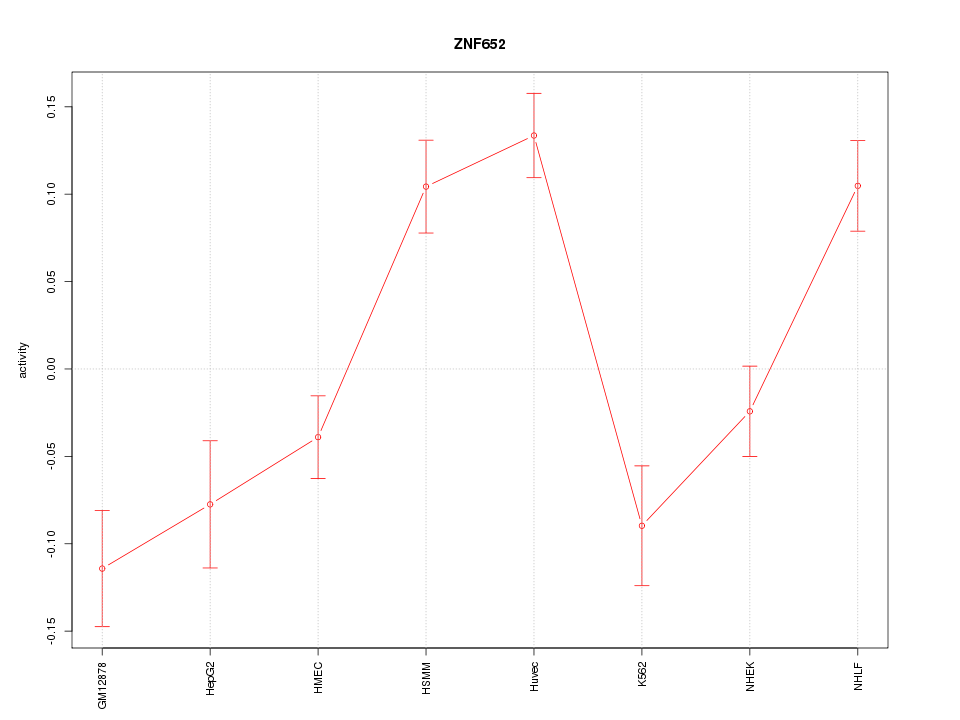
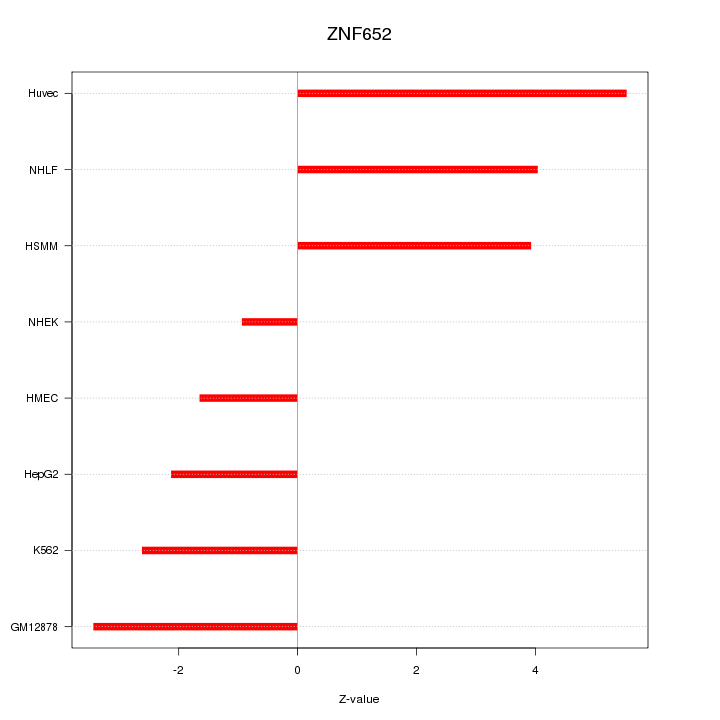
Motif ID: ZNF652
Z-value: 3.339
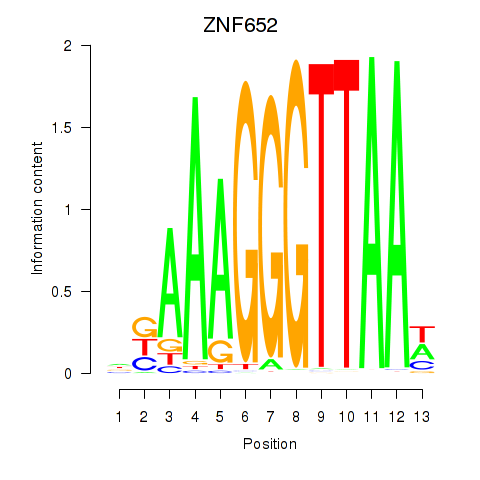
Transcription factors associated with ZNF652:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ZNF652 | ENSG00000198740.4 | ZNF652 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.6 | GO:0008228 | opsonization(GO:0008228) |
| 1.7 | 11.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.6 | 9.5 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 1.0 | 4.9 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.9 | 28.0 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.8 | 5.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.6 | 3.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.5 | 2.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 4.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 2.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 | 2.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 2.4 | GO:0022614 | membrane to membrane docking(GO:0022614) chorio-allantoic fusion(GO:0060710) |
| 0.4 | 2.3 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 4.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.3 | 7.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 1.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.2 | 3.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 1.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 0.5 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.2 | 1.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 1.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 2.3 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 2.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 1.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.4 | GO:0007028 | cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 3.7 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.3 | GO:2000831 | renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) obsolete regulation of natriuresis(GO:0003078) positive regulation of NAD(P)H oxidase activity(GO:0033864) corticosteroid hormone secretion(GO:0035930) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 | 10.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 2.0 | GO:0007220 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.8 | GO:0019934 | cGMP-mediated signaling(GO:0019934) regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.1 | 0.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.6 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 8.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.6 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.1 | 7.0 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.1 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.8 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.6 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.8 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.5 | GO:0006692 | prostanoid metabolic process(GO:0006692) |
| 0.0 | 1.9 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 1.4 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.8 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 2.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0071451 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 6.9 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.8 | GO:0072332 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 3.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.5 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 4.4 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.2 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.0 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 1.0 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 2.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.6 | 3.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 3.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 0.9 | GO:0043260 | laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.2 | 43.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.2 | 2.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 11.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0042589 | phagocytic vesicle membrane(GO:0030670) zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 5.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 10.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 5.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) protein acetyltransferase complex(GO:0031248) acetyltransferase complex(GO:1902493) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 23.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 3.1 | 28.0 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 1.8 | 5.4 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.5 | 11.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.1 | 4.3 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.8 | 9.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 3.8 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.7 | 3.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.6 | 4.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 1.8 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.4 | 2.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 5.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.4 | 2.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 0.4 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.3 | 0.9 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.3 | 1.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 5.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 5.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 1.7 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.1 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 7.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 3.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 7.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.0 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 2.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 4.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 5.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 4.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 4.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 3.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 3.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.7 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 6.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 2.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.2 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 3.6 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |