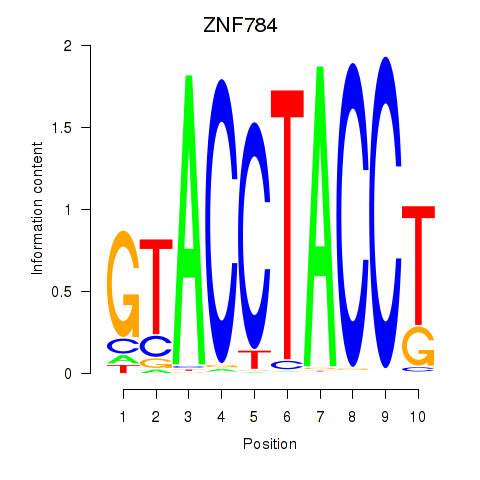
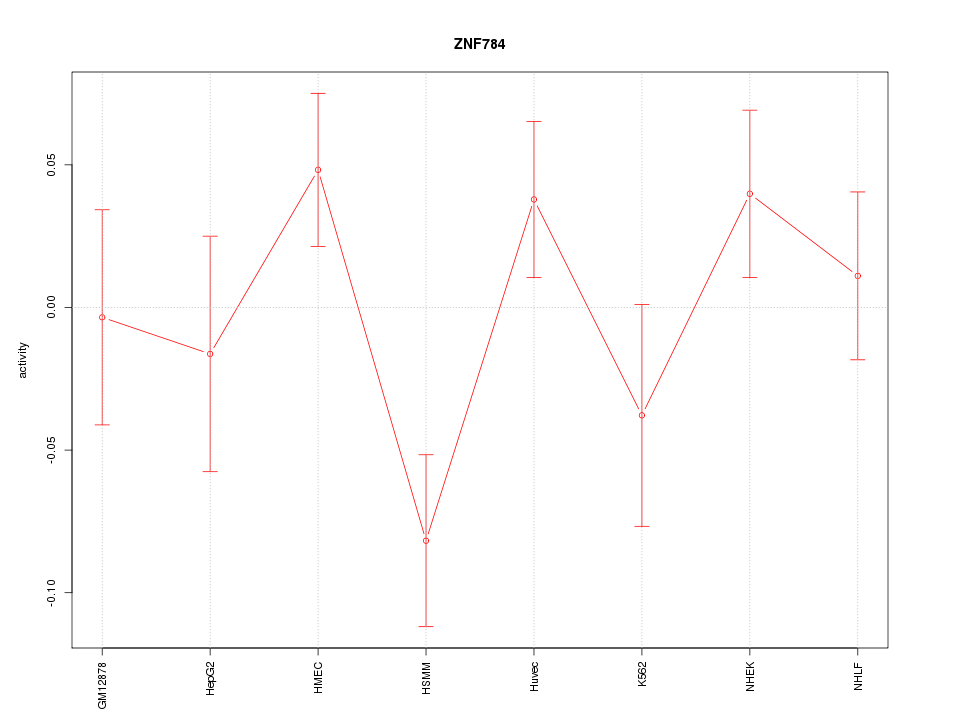
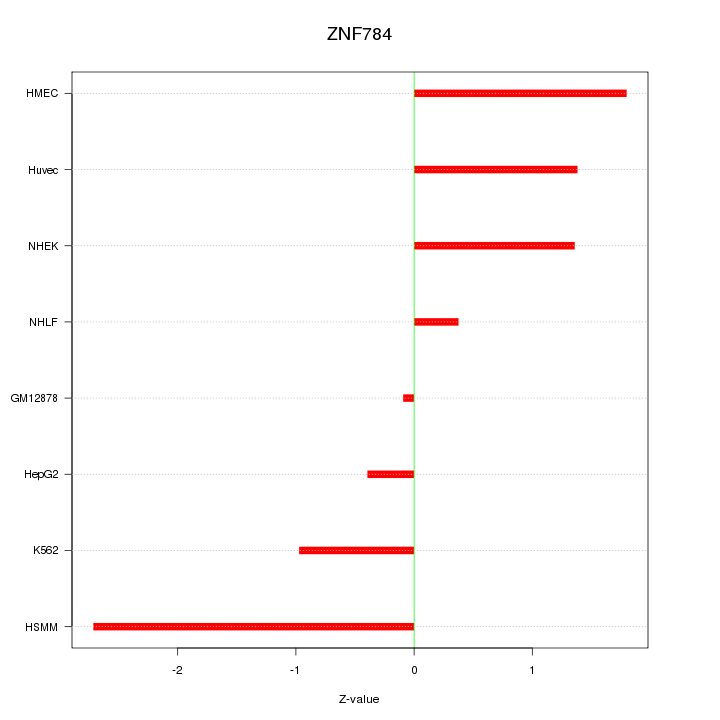
| 1.3 |
6.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 |
3.1 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.4 |
1.1 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.3 |
9.2 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.3 |
1.8 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 |
1.0 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 |
1.2 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.2 |
1.2 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.1 |
1.8 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
0.6 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.1 |
0.5 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.3 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.5 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.8 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
0.5 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.1 |
0.3 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.2 |
GO:0002331 |
immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 |
0.3 |
GO:0032099 |
negative regulation of appetite(GO:0032099) |
| 0.0 |
0.7 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 |
0.4 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.4 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.2 |
GO:0048550 |
regulation of pinocytosis(GO:0048548) negative regulation of pinocytosis(GO:0048550) |
| 0.0 |
0.2 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
0.0 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 |
0.2 |
GO:0060678 |
dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 |
0.8 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
2.4 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.5 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.1 |
GO:0071071 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.0 |
0.1 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 |
0.3 |
GO:0006525 |
arginine metabolic process(GO:0006525) |
| 0.0 |
0.2 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.6 |
GO:0042384 |
cilium assembly(GO:0042384) |
| 0.0 |
0.1 |
GO:0060122 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.4 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.2 |
GO:0008380 |
RNA splicing(GO:0008380) |
| 0.0 |
0.2 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.2 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
1.0 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.4 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
1.9 |
GO:0043627 |
response to estrogen(GO:0043627) |
| 0.0 |
0.3 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.1 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.9 |
GO:0009411 |
response to UV(GO:0009411) |
| 0.0 |
0.6 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.0 |
0.2 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |