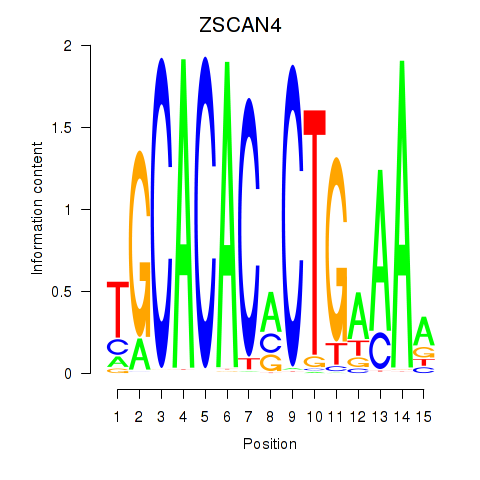
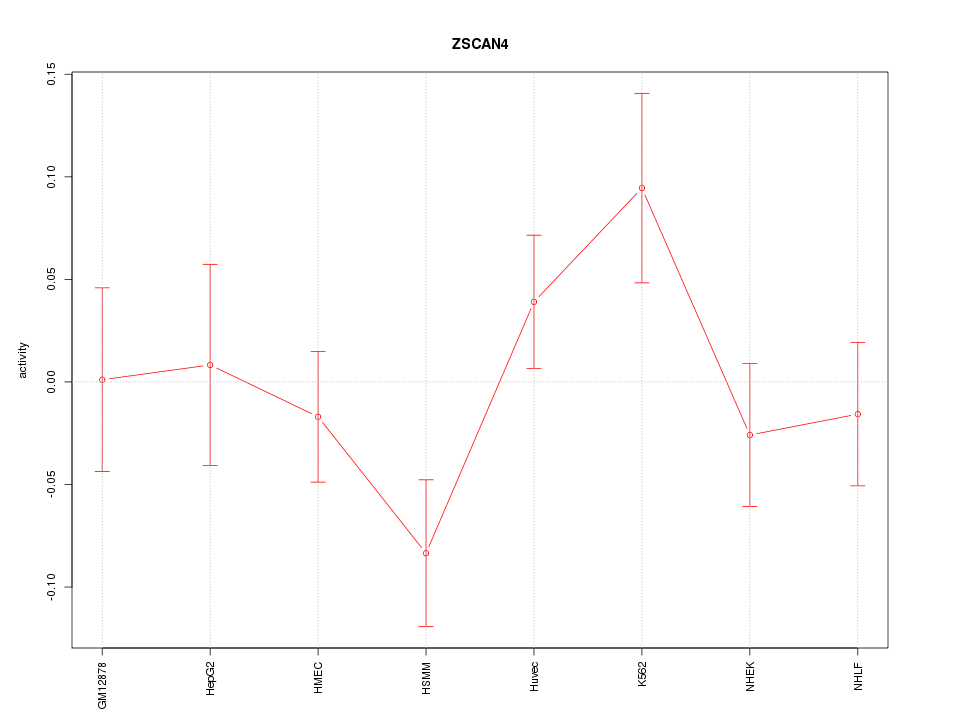
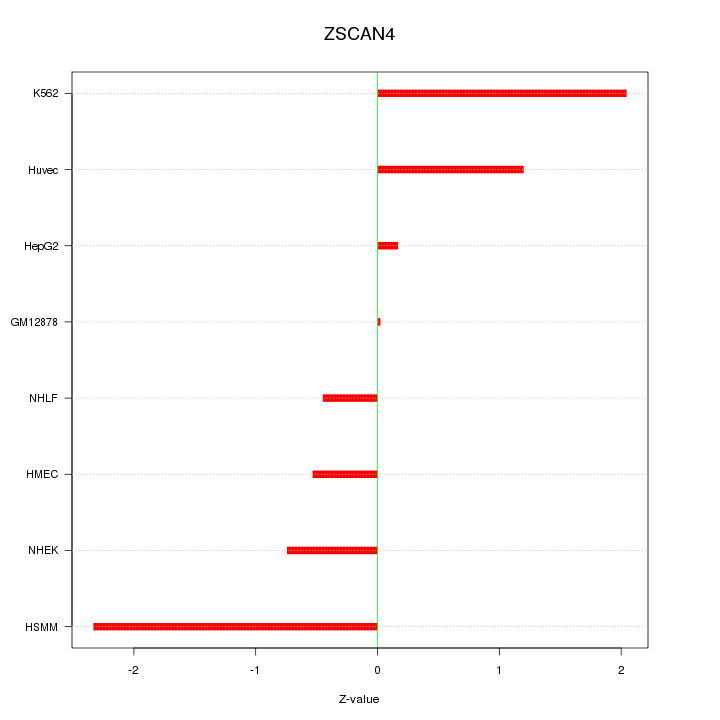
| 0.5 |
1.5 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.4 |
2.0 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 |
0.9 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.2 |
0.6 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.2 |
0.8 |
GO:0072526 |
pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 |
0.5 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.1 |
1.7 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.3 |
GO:0046984 |
regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 |
0.3 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 0.1 |
0.8 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.1 |
0.3 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 |
1.1 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 |
0.2 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) post-embryonic camera-type eye development(GO:0031077) |
| 0.0 |
1.2 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 |
0.2 |
GO:0014820 |
tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.9 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.3 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 |
0.4 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 |
0.1 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 |
0.1 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 |
0.1 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 |
0.4 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.1 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.0 |
0.1 |
GO:0051294 |
establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 |
0.1 |
GO:0042262 |
DNA protection(GO:0042262) |
| 0.0 |
0.2 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
2.5 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.7 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.3 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.9 |
GO:0007051 |
spindle organization(GO:0007051) |
| 0.0 |
0.2 |
GO:0040023 |
establishment of nucleus localization(GO:0040023) |
| 0.0 |
0.2 |
GO:0010458 |
exit from mitosis(GO:0010458) |