Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for AACACUG
Z-value: 0.69
miRNA associated with seed AACACUG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-141-3p
|
MIMAT0000432 |
|
hsa-miR-200a-3p
|
MIMAT0000682 |
Activity profile of AACACUG motif
Sorted Z-values of AACACUG motif
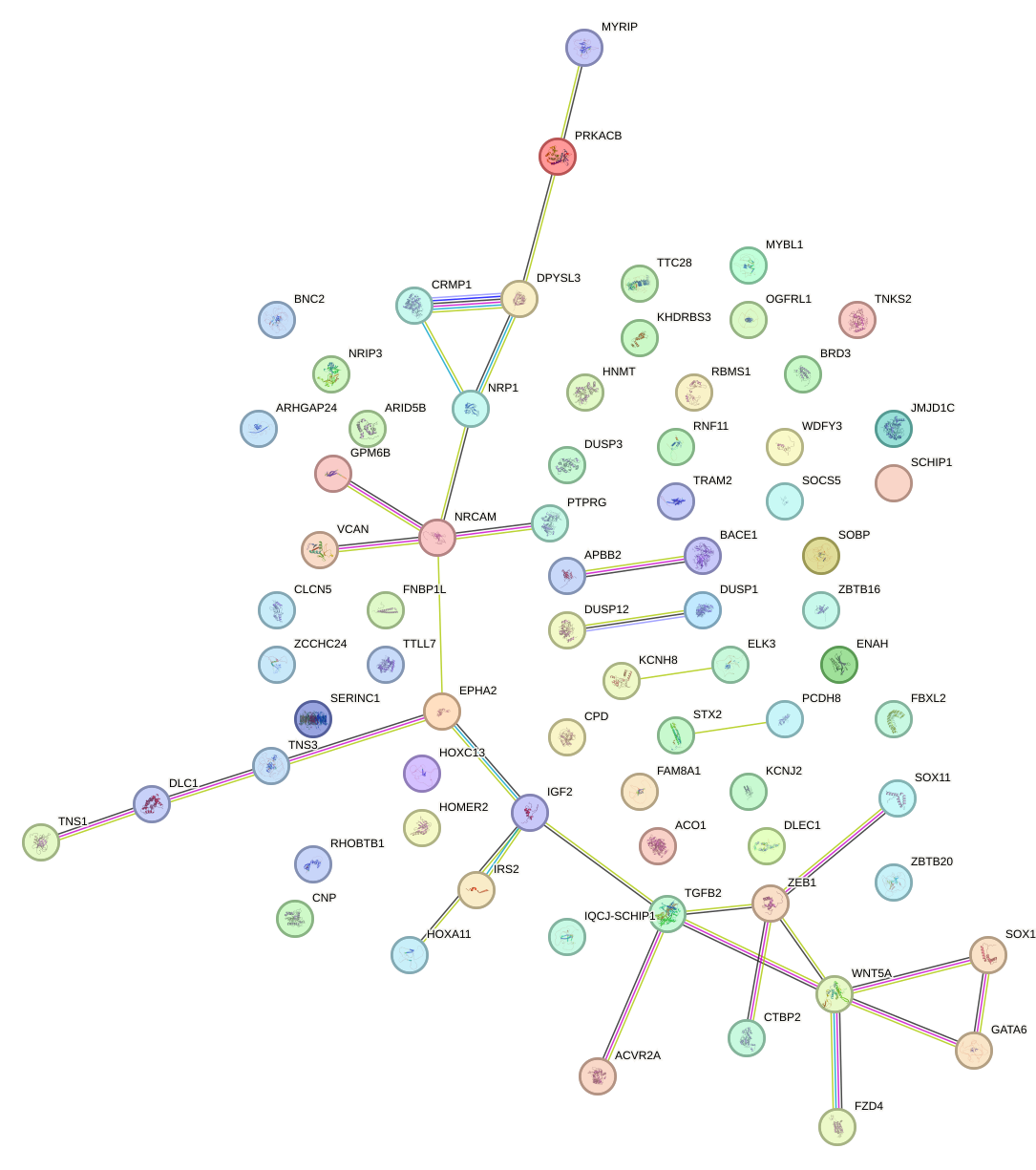
Network of associatons between targets according to the STRING database.
First level regulatory network of AACACUG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_47621736 | 1.67 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr11_-_2158507 | 1.59 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr10_+_31608054 | 1.46 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr3_+_61547585 | 1.09 |
ENST00000295874.10 ENST00000474889.1 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
| chr2_-_218808771 | 1.08 |
ENST00000449814.1 ENST00000171887.4 |
TNS1 |
tensin 1 |
| chr4_-_186877502 | 1.06 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr3_-_114790179 | 0.98 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_+_82767284 | 0.96 |
ENST00000265077.3 |
VCAN |
versican |
| chr22_-_29075853 | 0.93 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chrX_+_51928002 | 0.88 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr2_+_5832799 | 0.88 |
ENST00000322002.3 |
SOX11 |
SRY (sex determining region Y)-box 11 |
| chr10_-_81205373 | 0.87 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr10_-_62704005 | 0.84 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr14_-_30396948 | 0.84 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr9_+_32384617 | 0.84 |
ENST00000379923.1 ENST00000309951.6 ENST00000541043.1 |
ACO1 |
aconitase 1, soluble |
| chr4_+_86396265 | 0.82 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr8_-_67525473 | 0.81 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr3_+_159557637 | 0.80 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr3_+_158991025 | 0.74 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr6_-_52441713 | 0.73 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr2_-_161350305 | 0.67 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr6_+_107811162 | 0.67 |
ENST00000317357.5 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
| chr8_+_136469684 | 0.65 |
ENST00000355849.5 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_+_51701924 | 0.64 |
ENST00000242719.3 |
RNF11 |
ring finger protein 11 |
| chr4_-_111544254 | 0.63 |
ENST00000306732.3 |
PITX2 |
paired-like homeodomain 2 |
| chr3_+_62304712 | 0.62 |
ENST00000494481.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr3_+_187930719 | 0.59 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr1_+_93913713 | 0.58 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chr9_-_16870704 | 0.58 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr8_+_55370487 | 0.58 |
ENST00000297316.4 |
SOX17 |
SRY (sex determining region Y)-box 17 |
| chr1_+_84543734 | 0.56 |
ENST00000370689.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_+_68165657 | 0.55 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr10_-_33623564 | 0.55 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr2_+_102508955 | 0.54 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chr12_-_131323719 | 0.52 |
ENST00000392373.2 |
STX2 |
syntaxin 2 |
| chr17_-_8534067 | 0.51 |
ENST00000360416.3 ENST00000269243.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr6_+_71998506 | 0.51 |
ENST00000370435.4 |
OGFRL1 |
opioid growth factor receptor-like 1 |
| chr1_-_225840747 | 0.50 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr17_-_41856305 | 0.48 |
ENST00000397937.2 ENST00000226004.3 |
DUSP3 |
dual specificity phosphatase 3 |
| chr3_+_33318914 | 0.48 |
ENST00000484457.1 ENST00000538892.1 ENST00000538181.1 ENST00000446237.3 ENST00000507198.1 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
| chr4_-_129208940 | 0.47 |
ENST00000296425.5 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chrX_+_49687216 | 0.45 |
ENST00000376088.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr3_+_39851094 | 0.41 |
ENST00000302541.6 |
MYRIP |
myosin VIIA and Rab interacting protein |
| chr13_-_110438914 | 0.41 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr1_-_16482554 | 0.40 |
ENST00000358432.5 |
EPHA2 |
EPH receptor A2 |
| chrX_-_51812268 | 0.40 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr4_-_41216619 | 0.40 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr6_+_17600576 | 0.39 |
ENST00000259963.3 |
FAM8A1 |
family with sequence similarity 8, member A1 |
| chr8_-_13372395 | 0.38 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr9_-_23821273 | 0.37 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr11_-_86666427 | 0.37 |
ENST00000531380.1 |
FZD4 |
frizzled family receptor 4 |
| chr3_-_55523966 | 0.35 |
ENST00000474267.1 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr12_+_96588143 | 0.34 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr5_-_172198190 | 0.34 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr9_+_90112741 | 0.34 |
ENST00000469640.2 |
DAPK1 |
death-associated protein kinase 1 |
| chr4_-_85887503 | 0.33 |
ENST00000509172.1 ENST00000322366.6 ENST00000295888.4 ENST00000502713.1 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
| chr2_-_37899323 | 0.33 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chrX_-_13956737 | 0.32 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chr1_-_84464780 | 0.31 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr6_-_122792919 | 0.31 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chr7_-_27224795 | 0.30 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr1_+_218519577 | 0.29 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr3_-_185542817 | 0.29 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr11_-_117186946 | 0.29 |
ENST00000313005.6 ENST00000528053.1 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr18_-_53255766 | 0.29 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr14_-_61116168 | 0.28 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr9_-_136933134 | 0.28 |
ENST00000303407.7 |
BRD3 |
bromodomain containing 3 |
| chr2_+_148602058 | 0.28 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr10_-_126849068 | 0.27 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr7_-_111846435 | 0.25 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr9_-_74980113 | 0.24 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr4_-_5894777 | 0.24 |
ENST00000324989.7 |
CRMP1 |
collapsin response mediator protein 1 |
| chr3_+_171758344 | 0.23 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr18_+_19749386 | 0.21 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr10_+_63661053 | 0.21 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr2_+_138721850 | 0.21 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr10_+_93558069 | 0.20 |
ENST00000371627.4 |
TNKS2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr11_+_113930291 | 0.20 |
ENST00000335953.4 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
| chr2_+_46926048 | 0.20 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr17_+_66508537 | 0.20 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr17_+_28705921 | 0.19 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr2_+_30369807 | 0.19 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr10_-_65225722 | 0.19 |
ENST00000399251.1 |
JMJD1C |
jumonji domain containing 1C |
| chr11_-_9025541 | 0.18 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr7_-_108096822 | 0.18 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr12_+_54332535 | 0.18 |
ENST00000243056.3 |
HOXC13 |
homeobox C13 |
| chr13_-_53422640 | 0.17 |
ENST00000338862.4 ENST00000377942.3 |
PCDH8 |
protocadherin 8 |
| chr14_-_90085458 | 0.17 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr13_-_77460525 | 0.17 |
ENST00000377474.2 ENST00000317765.2 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
| chr17_+_40118759 | 0.16 |
ENST00000393892.3 |
CNP |
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_-_146833485 | 0.16 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr21_-_44846999 | 0.16 |
ENST00000270162.6 |
SIK1 |
salt-inducible kinase 1 |
| chr15_-_85259330 | 0.16 |
ENST00000560266.1 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
| chr5_-_132113036 | 0.16 |
ENST00000378706.1 |
SEPT8 |
septin 8 |
| chr3_+_43328004 | 0.15 |
ENST00000454177.1 ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK |
SNF related kinase |
| chr21_-_15755446 | 0.15 |
ENST00000544452.1 ENST00000285667.3 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
| chr3_+_132136331 | 0.15 |
ENST00000260818.6 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr17_-_46671323 | 0.15 |
ENST00000239151.5 |
HOXB5 |
homeobox B5 |
| chr9_-_36400213 | 0.15 |
ENST00000259605.6 ENST00000353739.4 |
RNF38 |
ring finger protein 38 |
| chr7_+_43152191 | 0.15 |
ENST00000395891.2 |
HECW1 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_128379346 | 0.15 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr16_+_53088885 | 0.15 |
ENST00000566029.1 ENST00000447540.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr15_+_57210818 | 0.15 |
ENST00000438423.2 ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12 |
transcription factor 12 |
| chr5_+_177019159 | 0.14 |
ENST00000332598.6 |
TMED9 |
transmembrane emp24 protein transport domain containing 9 |
| chr8_+_20054878 | 0.13 |
ENST00000276390.2 ENST00000519667.1 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
| chr2_-_240322643 | 0.13 |
ENST00000345617.3 |
HDAC4 |
histone deacetylase 4 |
| chr9_-_6015607 | 0.13 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr17_-_4167142 | 0.13 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr7_+_129710350 | 0.12 |
ENST00000335420.5 ENST00000463413.1 |
KLHDC10 |
kelch domain containing 10 |
| chr20_+_3776371 | 0.12 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr11_+_842808 | 0.11 |
ENST00000397397.2 ENST00000397411.2 ENST00000397396.1 |
TSPAN4 |
tetraspanin 4 |
| chr6_+_99282570 | 0.11 |
ENST00000328345.5 |
POU3F2 |
POU class 3 homeobox 2 |
| chr17_-_58603568 | 0.11 |
ENST00000083182.3 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr11_+_101981169 | 0.11 |
ENST00000526343.1 ENST00000282441.5 ENST00000537274.1 ENST00000345877.2 |
YAP1 |
Yes-associated protein 1 |
| chr20_+_3451650 | 0.11 |
ENST00000262919.5 |
ATRN |
attractin |
| chr5_-_95158375 | 0.11 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr17_-_74099795 | 0.11 |
ENST00000406660.3 ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7 |
exocyst complex component 7 |
| chr6_+_32121218 | 0.11 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_+_170102210 | 0.11 |
ENST00000439249.1 ENST00000332290.2 |
C6orf120 |
chromosome 6 open reading frame 120 |
| chr10_+_69644404 | 0.11 |
ENST00000212015.6 |
SIRT1 |
sirtuin 1 |
| chr17_-_46178527 | 0.11 |
ENST00000393408.3 |
CBX1 |
chromobox homolog 1 |
| chr16_-_71758602 | 0.10 |
ENST00000568954.1 |
PHLPP2 |
PH domain and leucine rich repeat protein phosphatase 2 |
| chr1_-_243418344 | 0.10 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr1_-_205719295 | 0.10 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr11_+_117049445 | 0.10 |
ENST00000324225.4 ENST00000532960.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr3_-_88108192 | 0.10 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chrX_+_21857717 | 0.10 |
ENST00000379484.5 |
MBTPS2 |
membrane-bound transcription factor peptidase, site 2 |
| chr5_-_32444828 | 0.10 |
ENST00000265069.8 |
ZFR |
zinc finger RNA binding protein |
| chr19_-_57352064 | 0.10 |
ENST00000326441.9 ENST00000593695.1 ENST00000599577.1 ENST00000594389.1 ENST00000423103.2 ENST00000598410.1 ENST00000593711.1 ENST00000391708.3 ENST00000221722.5 ENST00000599935.1 |
PEG3 ZIM2 |
paternally expressed 3 zinc finger, imprinted 2 |
| chr3_-_136471204 | 0.10 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr16_+_3507985 | 0.10 |
ENST00000421765.3 ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60 NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr5_+_145826867 | 0.10 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr1_+_114472222 | 0.09 |
ENST00000369558.1 ENST00000369561.4 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr12_-_16761007 | 0.09 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr11_+_33278811 | 0.09 |
ENST00000303296.4 ENST00000379016.3 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr3_+_11314099 | 0.09 |
ENST00000446450.2 ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7 |
autophagy related 7 |
| chr17_-_62502639 | 0.09 |
ENST00000225792.5 ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_+_244214577 | 0.09 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr11_-_73309228 | 0.09 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr22_+_38201114 | 0.08 |
ENST00000340857.2 |
H1F0 |
H1 histone family, member 0 |
| chr14_-_103523745 | 0.08 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr20_-_22565101 | 0.08 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr12_-_76478686 | 0.08 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr7_+_99613195 | 0.08 |
ENST00000324306.6 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr17_-_202579 | 0.08 |
ENST00000577079.1 ENST00000331302.7 ENST00000536489.2 |
RPH3AL |
rabphilin 3A-like (without C2 domains) |
| chr6_+_53659746 | 0.08 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr6_+_163835669 | 0.08 |
ENST00000453779.2 ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI |
QKI, KH domain containing, RNA binding |
| chr5_+_169010638 | 0.08 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr9_-_135819987 | 0.08 |
ENST00000298552.3 ENST00000403810.1 |
TSC1 |
tuberous sclerosis 1 |
| chr7_-_151217001 | 0.08 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr21_+_34697209 | 0.07 |
ENST00000270139.3 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
| chrX_-_119694538 | 0.07 |
ENST00000371322.5 |
CUL4B |
cullin 4B |
| chr20_-_4982132 | 0.07 |
ENST00000338244.1 ENST00000424750.2 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_-_173886491 | 0.07 |
ENST00000367698.3 |
SERPINC1 |
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr5_+_112312416 | 0.07 |
ENST00000389063.2 |
DCP2 |
decapping mRNA 2 |
| chr17_+_57642886 | 0.07 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr9_+_130374537 | 0.07 |
ENST00000373302.3 ENST00000373299.1 |
STXBP1 |
syntaxin binding protein 1 |
| chr2_+_178257372 | 0.07 |
ENST00000264167.4 ENST00000409888.1 |
AGPS |
alkylglycerone phosphate synthase |
| chr12_+_12938541 | 0.07 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr17_+_38171614 | 0.06 |
ENST00000583218.1 ENST00000394149.3 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
| chr9_-_123476719 | 0.06 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr2_-_50574856 | 0.06 |
ENST00000342183.5 |
NRXN1 |
neurexin 1 |
| chrX_+_153686614 | 0.06 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr5_-_148930960 | 0.06 |
ENST00000261798.5 ENST00000377843.2 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chr21_-_39870339 | 0.06 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr2_+_135676381 | 0.06 |
ENST00000537343.1 ENST00000295238.6 ENST00000264157.5 |
CCNT2 |
cyclin T2 |
| chr4_+_183164574 | 0.06 |
ENST00000511685.1 |
TENM3 |
teneurin transmembrane protein 3 |
| chr19_+_7459998 | 0.06 |
ENST00000319670.9 ENST00000599752.1 |
ARHGEF18 |
Rho/Rac guanine nucleotide exchange factor (GEF) 18 |
| chr1_+_110162448 | 0.06 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr3_-_116164306 | 0.06 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr15_+_77712993 | 0.05 |
ENST00000336216.4 ENST00000381714.3 ENST00000558651.1 |
HMG20A |
high mobility group 20A |
| chr15_+_62853562 | 0.05 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr19_+_19322758 | 0.05 |
ENST00000252575.6 |
NCAN |
neurocan |
| chr21_+_38071430 | 0.05 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr18_+_29171689 | 0.05 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr2_-_100939195 | 0.05 |
ENST00000393437.3 |
LONRF2 |
LON peptidase N-terminal domain and ring finger 2 |
| chr17_-_17875688 | 0.05 |
ENST00000379504.3 ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2 |
target of myb1-like 2 (chicken) |
| chrX_+_118370211 | 0.05 |
ENST00000217971.7 |
PGRMC1 |
progesterone receptor membrane component 1 |
| chr16_+_21610797 | 0.05 |
ENST00000358154.3 |
METTL9 |
methyltransferase like 9 |
| chr18_+_32558208 | 0.05 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chrX_+_40944871 | 0.05 |
ENST00000378308.2 ENST00000324545.8 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
| chr9_+_110045537 | 0.05 |
ENST00000358015.3 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
| chr13_-_73356009 | 0.04 |
ENST00000377780.4 ENST00000377767.4 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
| chr12_-_54673871 | 0.04 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr3_-_58419537 | 0.04 |
ENST00000474765.1 ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB |
pyruvate dehydrogenase (lipoamide) beta |
| chr4_+_699537 | 0.04 |
ENST00000419774.1 ENST00000362003.5 ENST00000400151.2 ENST00000427463.1 ENST00000470161.2 |
PCGF3 |
polycomb group ring finger 3 |
| chr16_-_30798492 | 0.04 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chr1_+_36621529 | 0.04 |
ENST00000316156.4 |
MAP7D1 |
MAP7 domain containing 1 |
| chr2_+_113403434 | 0.04 |
ENST00000272542.3 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
| chr10_+_111767720 | 0.04 |
ENST00000356080.4 ENST00000277900.8 |
ADD3 |
adducin 3 (gamma) |
| chr1_+_24742264 | 0.04 |
ENST00000374399.4 ENST00000003912.3 ENST00000358028.4 ENST00000339255.2 |
NIPAL3 |
NIPA-like domain containing 3 |
| chr22_+_46731596 | 0.04 |
ENST00000381019.3 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr2_+_71558858 | 0.04 |
ENST00000437658.2 ENST00000355812.3 ENST00000377802.2 ENST00000264447.4 ENST00000409544.1 ENST00000455226.1 ENST00000454278.1 ENST00000417778.1 ENST00000454122.1 |
ZNF638 |
zinc finger protein 638 |
| chr2_-_167232484 | 0.04 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr9_+_112810878 | 0.04 |
ENST00000434623.2 ENST00000374525.1 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
| chr10_+_76586348 | 0.04 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr1_+_118148556 | 0.04 |
ENST00000369448.3 |
FAM46C |
family with sequence similarity 46, member C |
| chr2_-_131850951 | 0.04 |
ENST00000409185.1 ENST00000389915.3 |
FAM168B |
family with sequence similarity 168, member B |
| chr1_+_40723779 | 0.04 |
ENST00000372759.3 |
ZMPSTE24 |
zinc metallopeptidase STE24 |
| chr11_+_36589547 | 0.03 |
ENST00000299440.5 |
RAG1 |
recombination activating gene 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 1.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.5 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 1.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 1.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.1 | 1.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.6 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.5 | GO:0038189 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.8 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.7 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.3 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.2 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 0.1 | 0.4 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.3 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.3 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0051138 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 1.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.7 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1901859 | late nucleophagy(GO:0044805) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0035822 | release from viral latency(GO:0019046) gene conversion(GO:0035822) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.0 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.0 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.6 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.4 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 3.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |