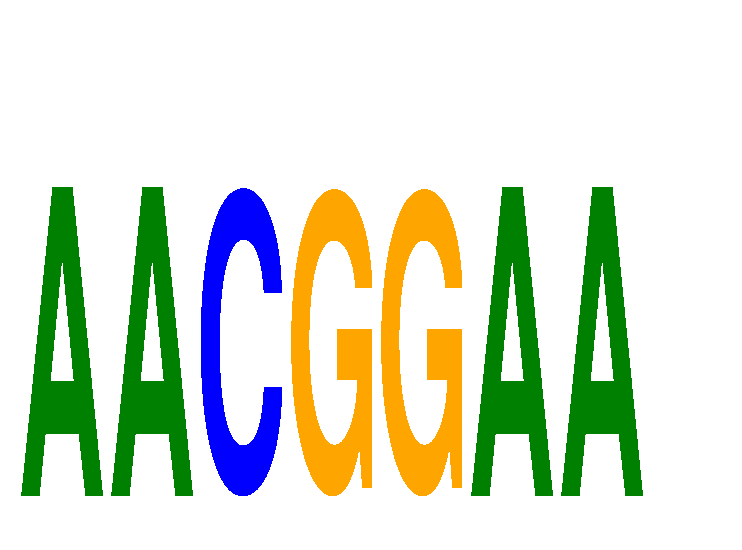Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for AACGGAA
Z-value: 0.14
miRNA associated with seed AACGGAA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-191-5p
|
MIMAT0000440 |
Activity profile of AACGGAA motif
Sorted Z-values of AACGGAA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AACGGAA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_46623441 | 0.28 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chrX_-_109561294 | 0.16 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr10_-_81205373 | 0.11 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr19_+_3359561 | 0.06 |
ENST00000589123.1 ENST00000346156.5 ENST00000395111.3 ENST00000586919.1 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr19_-_15560730 | 0.05 |
ENST00000389282.4 ENST00000263381.7 |
WIZ |
widely interspaced zinc finger motifs |
| chr7_-_108096822 | 0.05 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr5_+_149887672 | 0.05 |
ENST00000261797.6 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr6_+_116601265 | 0.04 |
ENST00000452085.3 |
DSE |
dermatan sulfate epimerase |
| chr7_-_92463210 | 0.04 |
ENST00000265734.4 |
CDK6 |
cyclin-dependent kinase 6 |
| chr9_-_16870704 | 0.03 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr9_+_133454943 | 0.02 |
ENST00000319725.9 |
FUBP3 |
far upstream element (FUSE) binding protein 3 |
| chr12_+_122242597 | 0.02 |
ENST00000267197.5 |
SETD1B |
SET domain containing 1B |
| chr3_-_18466787 | 0.02 |
ENST00000338745.6 ENST00000450898.1 |
SATB1 |
SATB homeobox 1 |
| chr3_-_52090461 | 0.01 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr7_-_17980091 | 0.01 |
ENST00000409389.1 ENST00000409604.1 ENST00000428135.3 |
SNX13 |
sorting nexin 13 |
| chr16_-_4897266 | 0.01 |
ENST00000591451.1 ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr12_-_58240470 | 0.00 |
ENST00000548823.1 ENST00000398073.2 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr8_-_134115118 | 0.00 |
ENST00000395352.3 ENST00000338087.5 |
SLA |
Src-like-adaptor |
| chr9_-_3525968 | 0.00 |
ENST00000382004.3 ENST00000302303.1 ENST00000449190.1 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
| chr15_+_52043758 | 0.00 |
ENST00000249700.4 ENST00000539962.2 |
TMOD2 |
tropomodulin 2 (neuronal) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021569 | rhombomere 3 development(GO:0021569) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.0 | GO:0098770 | FBXO family protein binding(GO:0098770) |