Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ACAGUAU
Z-value: 1.15
miRNA associated with seed ACAGUAU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-144-3p
|
MIMAT0000436 |
Activity profile of ACAGUAU motif
Sorted Z-values of ACAGUAU motif
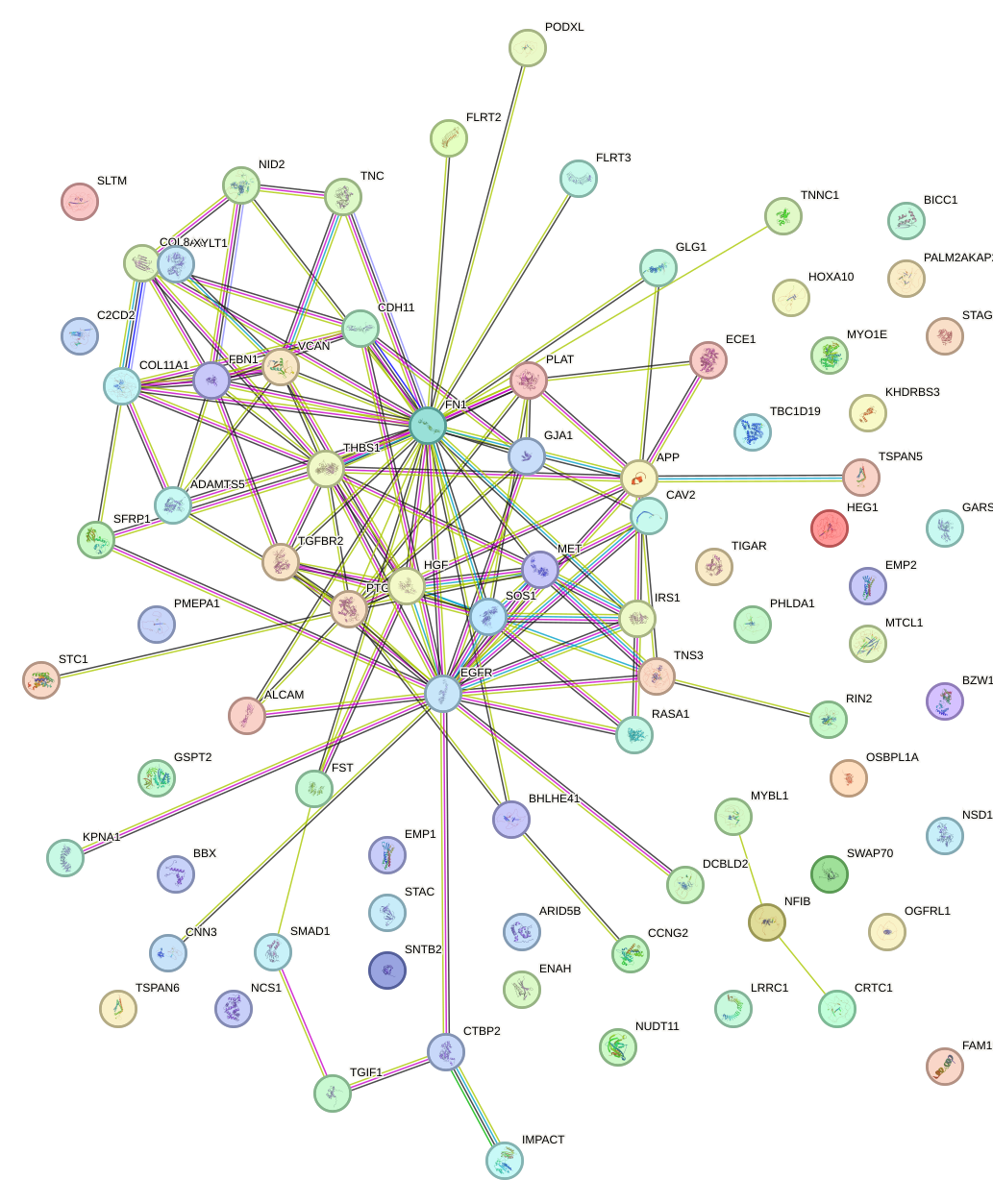
Network of associatons between targets according to the STRING database.
First level regulatory network of ACAGUAU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_52776228 | 5.42 |
ENST00000256759.3 |
FST |
follistatin |
| chrX_+_102631248 | 4.94 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr8_-_41166953 | 4.93 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr16_-_65155833 | 4.02 |
ENST00000566827.1 ENST00000394156.3 ENST00000562998.1 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_-_95392635 | 3.58 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chr15_+_39873268 | 3.55 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr9_-_117880477 | 2.91 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr2_-_227664474 | 2.89 |
ENST00000305123.5 |
IRS1 |
insulin receptor substrate 1 |
| chr12_-_15942309 | 2.77 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr5_-_127873659 | 2.74 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr3_+_30648066 | 2.29 |
ENST00000359013.4 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
| chr6_+_121756809 | 2.25 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr8_+_136469684 | 2.00 |
ENST00000355849.5 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
| chr4_-_7873981 | 1.89 |
ENST00000360265.4 |
AFAP1 |
actin filament associated protein 1 |
| chr10_+_60272814 | 1.88 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr20_-_14318248 | 1.84 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr4_-_99579733 | 1.83 |
ENST00000305798.3 |
TSPAN5 |
tetraspanin 5 |
| chr21_-_27542972 | 1.83 |
ENST00000346798.3 ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP |
amyloid beta (A4) precursor protein |
| chr10_-_126849068 | 1.83 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr14_-_52535712 | 1.67 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr3_-_98620500 | 1.58 |
ENST00000326840.6 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
| chr6_+_114178512 | 1.55 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr9_+_132934835 | 1.49 |
ENST00000372398.3 |
NCS1 |
neuronal calcium sensor 1 |
| chr13_-_107187462 | 1.45 |
ENST00000245323.4 |
EFNB2 |
ephrin-B2 |
| chr12_-_76425368 | 1.42 |
ENST00000602540.1 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
| chr15_-_48937982 | 1.41 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr14_+_85996471 | 1.36 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr16_-_10674528 | 1.35 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr20_+_19867150 | 1.27 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr1_-_225840747 | 1.27 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr8_-_42397037 | 1.25 |
ENST00000342228.3 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
| chr12_+_13349650 | 1.24 |
ENST00000256951.5 ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1 |
epithelial membrane protein 1 |
| chr3_+_36421826 | 1.18 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr11_-_115375107 | 1.16 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr10_+_63661053 | 1.15 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr12_-_26278030 | 1.14 |
ENST00000242728.4 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
| chr8_-_42065187 | 1.13 |
ENST00000270189.6 ENST00000352041.3 ENST00000220809.4 |
PLAT |
plasminogen activator, tissue |
| chrX_-_99891796 | 1.09 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr12_+_4430371 | 1.08 |
ENST00000179259.4 |
C12orf5 |
chromosome 12 open reading frame 5 |
| chr3_+_99357319 | 1.02 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr21_-_43346790 | 1.02 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr18_+_8717369 | 1.01 |
ENST00000359865.3 ENST00000400050.3 ENST00000306285.7 |
SOGA2 |
SOGA family member 2 |
| chr7_+_116139424 | 0.99 |
ENST00000222693.4 |
CAV2 |
caveolin 2 |
| chr2_-_37899323 | 0.96 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_-_235405679 | 0.92 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr3_+_105085734 | 0.92 |
ENST00000306107.5 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr7_+_55086794 | 0.89 |
ENST00000275493.2 ENST00000442591.1 |
EGFR |
epidermal growth factor receptor |
| chr13_-_77460525 | 0.88 |
ENST00000377474.2 ENST00000317765.2 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
| chr7_-_27213893 | 0.88 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr1_-_186649543 | 0.87 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr16_+_69221028 | 0.86 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr3_-_124774802 | 0.82 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr6_-_139695757 | 0.82 |
ENST00000367651.2 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr4_+_78078304 | 0.82 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
| chr5_+_82767284 | 0.80 |
ENST00000265077.3 |
VCAN |
versican |
| chr7_-_47621736 | 0.77 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr19_+_18794470 | 0.76 |
ENST00000321949.8 ENST00000338797.6 |
CRTC1 |
CREB regulated transcription coactivator 1 |
| chr9_+_112810878 | 0.74 |
ENST00000434623.2 ENST00000374525.1 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
| chr3_+_107241783 | 0.74 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr18_+_22006580 | 0.73 |
ENST00000284202.4 |
IMPACT |
impact RWD domain protein |
| chrX_+_51486481 | 0.70 |
ENST00000340438.4 |
GSPT2 |
G1 to S phase transition 2 |
| chr6_+_53659746 | 0.69 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr5_-_38595498 | 0.69 |
ENST00000263409.4 |
LIFR |
leukemia inhibitory factor receptor alpha |
| chr3_+_51428704 | 0.68 |
ENST00000323686.4 |
RBM15B |
RNA binding motif protein 15B |
| chr7_-_81399438 | 0.67 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_134210243 | 0.67 |
ENST00000367882.4 |
TCF21 |
transcription factor 21 |
| chr15_-_59665062 | 0.66 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr18_+_3451646 | 0.65 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr9_+_112542572 | 0.65 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr21_-_39288743 | 0.65 |
ENST00000609713.1 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr8_-_67525473 | 0.63 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr5_-_176981417 | 0.63 |
ENST00000514747.1 ENST00000443375.2 ENST00000329540.5 |
FAM193B |
family with sequence similarity 193, member B |
| chr16_-_74640986 | 0.61 |
ENST00000422840.2 ENST00000565260.1 ENST00000447066.2 ENST00000205061.5 |
GLG1 |
golgi glycoprotein 1 |
| chr8_-_23712312 | 0.60 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chr20_+_11871371 | 0.59 |
ENST00000254977.3 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr2_-_216300784 | 0.58 |
ENST00000421182.1 ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1 |
fibronectin 1 |
| chr6_+_71998506 | 0.58 |
ENST00000370435.4 |
OGFRL1 |
opioid growth factor receptor-like 1 |
| chr9_-_14314066 | 0.57 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr4_+_146402925 | 0.56 |
ENST00000302085.4 |
SMAD1 |
SMAD family member 1 |
| chr3_-_136471204 | 0.56 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr7_-_131241361 | 0.55 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr11_+_9685604 | 0.55 |
ENST00000447399.2 ENST00000318950.6 |
SWAP70 |
SWAP switching B-cell complex 70kDa subunit |
| chr18_-_21977748 | 0.55 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr5_+_86564739 | 0.55 |
ENST00000456692.2 ENST00000512763.1 ENST00000506290.1 |
RASA1 |
RAS p21 protein activator (GTPase activating protein) 1 |
| chr21_-_28338732 | 0.55 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chrX_-_51239425 | 0.54 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr16_-_17564738 | 0.54 |
ENST00000261381.6 |
XYLT1 |
xylosyltransferase I |
| chr2_+_201676256 | 0.53 |
ENST00000452206.1 ENST00000410110.2 ENST00000409600.1 |
BZW1 |
basic leucine zipper and W2 domains 1 |
| chr8_+_98881268 | 0.53 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr1_-_103574024 | 0.52 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr3_-_122233723 | 0.51 |
ENST00000493510.1 ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1 |
karyopherin alpha 1 (importin alpha 5) |
| chr4_+_26585538 | 0.51 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr5_+_176560742 | 0.49 |
ENST00000439151.2 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr1_-_21671968 | 0.47 |
ENST00000415912.2 |
ECE1 |
endothelin converting enzyme 1 |
| chr7_+_116312411 | 0.47 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr17_-_78450398 | 0.47 |
ENST00000306773.4 |
NPTX1 |
neuronal pentraxin I |
| chr16_+_86544113 | 0.46 |
ENST00000262426.4 |
FOXF1 |
forkhead box F1 |
| chr10_+_60094735 | 0.46 |
ENST00000373910.4 |
UBE2D1 |
ubiquitin-conjugating enzyme E2D 1 |
| chr7_-_6312206 | 0.45 |
ENST00000350796.3 |
CYTH3 |
cytohesin 3 |
| chr10_-_79686284 | 0.44 |
ENST00000372391.2 ENST00000372388.2 |
DLG5 |
discs, large homolog 5 (Drosophila) |
| chr12_+_27485823 | 0.44 |
ENST00000395901.2 ENST00000546179.1 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr5_-_172198190 | 0.44 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr16_-_47177874 | 0.43 |
ENST00000562435.1 |
NETO2 |
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr2_+_5832799 | 0.43 |
ENST00000322002.3 |
SOX11 |
SRY (sex determining region Y)-box 11 |
| chr9_-_23821273 | 0.41 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_141488070 | 0.40 |
ENST00000253814.4 |
NDFIP1 |
Nedd4 family interacting protein 1 |
| chr2_+_198380289 | 0.40 |
ENST00000233892.4 ENST00000409916.1 |
MOB4 |
MOB family member 4, phocein |
| chr12_-_57505121 | 0.40 |
ENST00000538913.2 ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr15_-_83876758 | 0.40 |
ENST00000299633.4 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
| chr6_+_148663729 | 0.39 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr7_-_27196267 | 0.39 |
ENST00000242159.3 |
HOXA7 |
homeobox A7 |
| chr9_+_77112244 | 0.39 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr14_-_81687197 | 0.39 |
ENST00000553612.1 |
GTF2A1 |
general transcription factor IIA, 1, 19/37kDa |
| chr9_-_16870704 | 0.38 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr8_+_79578282 | 0.37 |
ENST00000263849.4 |
ZC2HC1A |
zinc finger, C2HC-type containing 1A |
| chr4_-_139163491 | 0.36 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr1_+_87170247 | 0.36 |
ENST00000370558.4 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
| chr4_+_126237554 | 0.36 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chr4_+_183164574 | 0.36 |
ENST00000511685.1 |
TENM3 |
teneurin transmembrane protein 3 |
| chr18_-_61089665 | 0.36 |
ENST00000238497.5 |
VPS4B |
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr6_+_143929307 | 0.36 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr19_+_45973120 | 0.36 |
ENST00000592811.1 ENST00000586615.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr15_+_96873921 | 0.34 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_200708671 | 0.34 |
ENST00000358823.2 |
CAMSAP2 |
calmodulin regulated spectrin-associated protein family, member 2 |
| chrX_-_108976521 | 0.34 |
ENST00000469796.2 ENST00000502391.1 ENST00000508092.1 ENST00000340800.2 ENST00000348502.6 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
| chr2_-_161056802 | 0.33 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr5_-_95297678 | 0.33 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr10_+_8096631 | 0.33 |
ENST00000379328.3 |
GATA3 |
GATA binding protein 3 |
| chr11_-_27494279 | 0.32 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr9_+_128509624 | 0.32 |
ENST00000342287.5 ENST00000373487.4 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr11_+_129939779 | 0.32 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr1_+_36621529 | 0.32 |
ENST00000316156.4 |
MAP7D1 |
MAP7 domain containing 1 |
| chr15_-_30114622 | 0.31 |
ENST00000495972.2 ENST00000346128.6 |
TJP1 |
tight junction protein 1 |
| chr1_+_186798073 | 0.31 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_-_18691739 | 0.31 |
ENST00000399799.2 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
| chr21_-_36260980 | 0.31 |
ENST00000344691.4 ENST00000358356.5 |
RUNX1 |
runt-related transcription factor 1 |
| chr15_-_34502278 | 0.31 |
ENST00000559515.1 ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr10_-_3827417 | 0.31 |
ENST00000497571.1 ENST00000542957.1 |
KLF6 |
Kruppel-like factor 6 |
| chr15_-_56209306 | 0.31 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr10_-_735553 | 0.31 |
ENST00000280886.6 ENST00000423550.1 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_-_70719925 | 0.31 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr4_-_129208940 | 0.30 |
ENST00000296425.5 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr17_+_61086917 | 0.30 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_+_220701456 | 0.30 |
ENST00000366918.4 ENST00000402574.1 |
MARK1 |
MAP/microtubule affinity-regulating kinase 1 |
| chr6_-_82462425 | 0.30 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr1_+_198126093 | 0.29 |
ENST00000367385.4 ENST00000442588.1 ENST00000538004.1 |
NEK7 |
NIMA-related kinase 7 |
| chr14_+_65171099 | 0.28 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr16_+_8891670 | 0.28 |
ENST00000268261.4 ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2 |
phosphomannomutase 2 |
| chr14_-_82000140 | 0.28 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr8_-_53322303 | 0.28 |
ENST00000276480.7 |
ST18 |
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr11_+_69924397 | 0.28 |
ENST00000355303.5 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr5_-_132299313 | 0.27 |
ENST00000265343.5 |
AFF4 |
AF4/FMR2 family, member 4 |
| chrX_-_41782249 | 0.27 |
ENST00000442742.2 ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr12_-_28124903 | 0.27 |
ENST00000395872.1 ENST00000354417.3 ENST00000201015.4 |
PTHLH |
parathyroid hormone-like hormone |
| chr16_+_67927147 | 0.27 |
ENST00000291041.5 |
PSKH1 |
protein serine kinase H1 |
| chr3_-_99833333 | 0.27 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr3_-_119813264 | 0.26 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr12_-_51477333 | 0.26 |
ENST00000228515.1 ENST00000548206.1 ENST00000546935.1 ENST00000548981.1 |
CSRNP2 |
cysteine-serine-rich nuclear protein 2 |
| chr3_-_114790179 | 0.26 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr15_-_65809581 | 0.26 |
ENST00000341861.5 |
DPP8 |
dipeptidyl-peptidase 8 |
| chr6_+_107811162 | 0.25 |
ENST00000317357.5 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
| chr1_+_155829286 | 0.25 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr1_+_82266053 | 0.24 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr13_-_50367057 | 0.24 |
ENST00000261667.3 |
KPNA3 |
karyopherin alpha 3 (importin alpha 4) |
| chr14_+_45431379 | 0.24 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr13_+_49550015 | 0.24 |
ENST00000492622.2 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr20_-_61493115 | 0.23 |
ENST00000335351.3 ENST00000217162.5 |
TCFL5 |
transcription factor-like 5 (basic helix-loop-helix) |
| chr9_-_107690420 | 0.23 |
ENST00000423487.2 ENST00000374733.1 ENST00000374736.3 |
ABCA1 |
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr6_+_89790490 | 0.22 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr17_-_58469474 | 0.22 |
ENST00000300896.4 |
USP32 |
ubiquitin specific peptidase 32 |
| chr14_+_75745477 | 0.22 |
ENST00000303562.4 ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr6_-_8064567 | 0.22 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr17_-_4269768 | 0.21 |
ENST00000396981.2 |
UBE2G1 |
ubiquitin-conjugating enzyme E2G 1 |
| chr13_-_48575443 | 0.21 |
ENST00000378654.3 |
SUCLA2 |
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_-_53737795 | 0.21 |
ENST00000262135.4 ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L |
RPGRIP1-like |
| chr2_-_183903133 | 0.21 |
ENST00000361354.4 |
NCKAP1 |
NCK-associated protein 1 |
| chr1_-_38512450 | 0.21 |
ENST00000373012.2 |
POU3F1 |
POU class 3 homeobox 1 |
| chr15_+_77223960 | 0.21 |
ENST00000394885.3 |
RCN2 |
reticulocalbin 2, EF-hand calcium binding domain |
| chr3_+_150321068 | 0.20 |
ENST00000471696.1 ENST00000477889.1 ENST00000485923.1 |
SELT |
Selenoprotein T |
| chr4_+_72204755 | 0.20 |
ENST00000512686.1 ENST00000340595.3 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_-_54930790 | 0.20 |
ENST00000263921.3 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
| chr16_+_27561449 | 0.20 |
ENST00000261588.4 |
KIAA0556 |
KIAA0556 |
| chr2_-_178128528 | 0.19 |
ENST00000397063.4 ENST00000421929.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr1_-_78148324 | 0.19 |
ENST00000370801.3 ENST00000433749.1 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
| chr10_+_93558069 | 0.19 |
ENST00000371627.4 |
TNKS2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr3_-_116164306 | 0.19 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr16_-_19533404 | 0.19 |
ENST00000353258.3 |
GDE1 |
glycerophosphodiester phosphodiesterase 1 |
| chr3_+_130569429 | 0.19 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_-_132834184 | 0.19 |
ENST00000367941.2 ENST00000367937.4 |
STX7 |
syntaxin 7 |
| chr12_-_90049828 | 0.18 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_123476719 | 0.18 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr14_-_99737565 | 0.18 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr8_+_61429416 | 0.16 |
ENST00000262646.7 ENST00000531289.1 |
RAB2A |
RAB2A, member RAS oncogene family |
| chr10_+_96162242 | 0.16 |
ENST00000225235.4 |
TBC1D12 |
TBC1 domain family, member 12 |
| chr10_+_95256356 | 0.16 |
ENST00000371485.3 |
CEP55 |
centrosomal protein 55kDa |
| chr10_+_76586348 | 0.16 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr5_+_71403061 | 0.16 |
ENST00000512974.1 ENST00000296755.7 |
MAP1B |
microtubule-associated protein 1B |
| chr15_-_77363513 | 0.16 |
ENST00000267970.4 |
TSPAN3 |
tetraspanin 3 |
| chr7_-_120498357 | 0.15 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr6_-_56707943 | 0.15 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 10.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.9 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 4.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 4.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 7.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.6 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 4.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 0.9 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.3 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 4.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 2.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.5 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 1.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 1.8 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 5.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 1.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 2.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.9 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 6.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 2.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.2 | 3.6 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.8 | 2.3 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.8 | 2.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.7 | 2.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.7 | 2.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.7 | 5.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 1.4 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.5 | 4.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 1.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 4.0 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.4 | 1.8 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 0.9 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.3 | 1.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 | 2.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 0.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 2.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 1.7 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.2 | 0.7 | GO:0072277 | branchiomeric skeletal muscle development(GO:0014707) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.8 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 1.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.2 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 0.7 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.7 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 0.7 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 | 3.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 1.6 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 0.5 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.7 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 1.5 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.4 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.5 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 1.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.9 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.5 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 1.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.2 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 0.6 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0071896 | negative regulation of hippo signaling(GO:0035331) protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 1.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.2 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0046469 | corpus callosum morphogenesis(GO:0021540) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.2 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.8 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 1.1 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.9 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.5 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 1.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.8 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032455 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) renin secretion into blood stream(GO:0002001) nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.4 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.0 | GO:2000364 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.0 | GO:0072716 | response to actinomycin D(GO:0072716) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 1.6 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 1.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.9 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.3 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.6 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:1900449 | regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 2.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 4.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 2.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 1.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 2.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.8 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 4.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 0.7 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 0.9 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 3.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 4.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 0.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 3.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0070697 | activin receptor binding(GO:0070697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.0 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |