Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ACAUUCA
Z-value: 0.55
miRNA associated with seed ACAUUCA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-181a-5p
|
MIMAT0000256 |
|
hsa-miR-181b-5p
|
MIMAT0000257 |
|
hsa-miR-181c-5p
|
MIMAT0000258 |
|
hsa-miR-181d-5p
|
MIMAT0002821 |
|
hsa-miR-4262
|
MIMAT0016894 |
Activity profile of ACAUUCA motif
Sorted Z-values of ACAUUCA motif
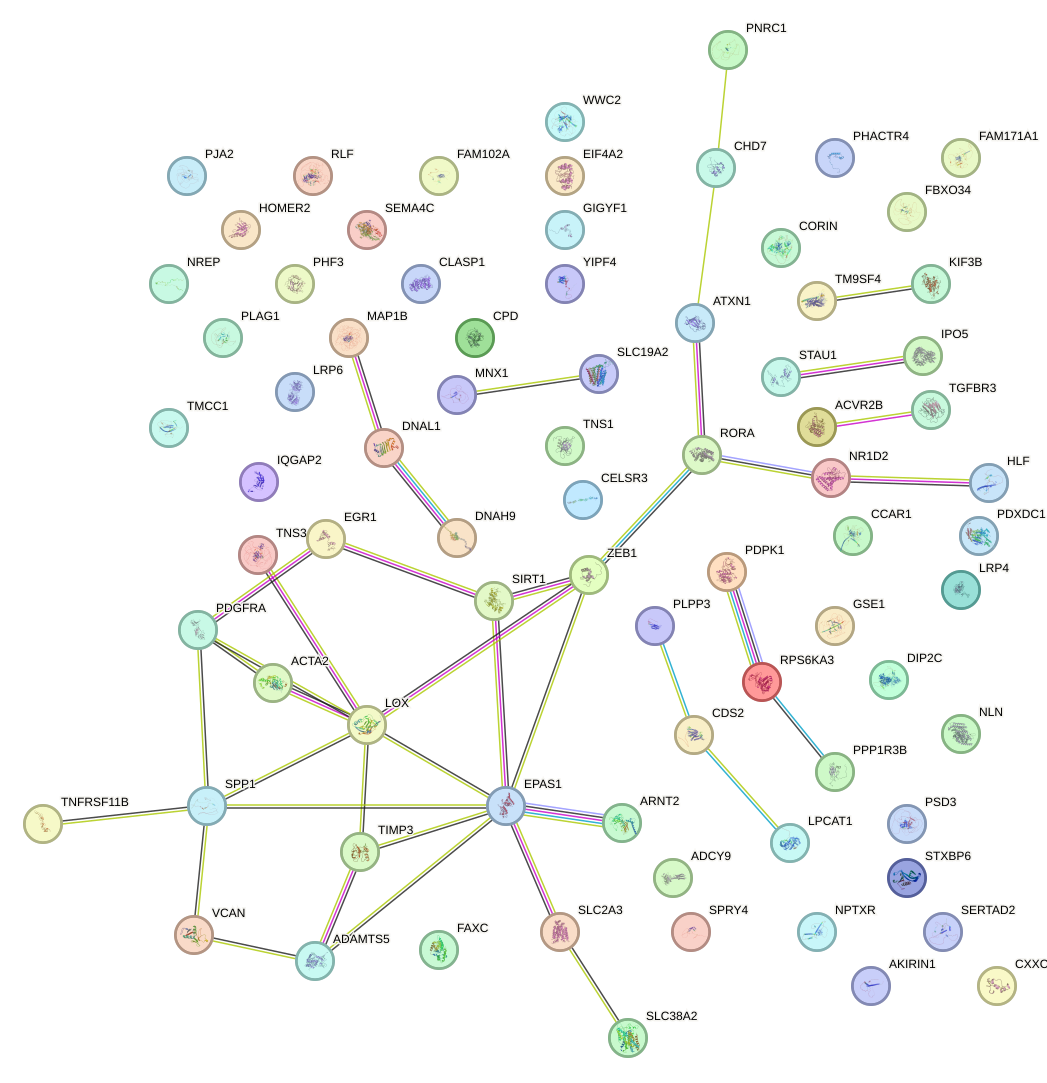
Network of associatons between targets according to the STRING database.
First level regulatory network of ACAUUCA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_15016725 | 1.04 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr17_+_53342311 | 0.91 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr7_-_47621736 | 0.84 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr1_-_92351769 | 0.84 |
ENST00000212355.4 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr1_-_57045228 | 0.77 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr22_+_33197683 | 0.67 |
ENST00000266085.6 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
| chr16_+_85646763 | 0.67 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr2_-_218808771 | 0.66 |
ENST00000449814.1 ENST00000171887.4 |
TNS1 |
tensin 1 |
| chr14_-_25519095 | 0.61 |
ENST00000419632.2 ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr1_-_169455169 | 0.61 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr1_+_214161272 | 0.59 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr16_+_69599861 | 0.59 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr2_-_64881018 | 0.58 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr15_-_61521495 | 0.57 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr5_-_121413974 | 0.57 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr3_+_38495333 | 0.55 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr5_+_75699040 | 0.54 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr16_-_4166186 | 0.52 |
ENST00000294016.3 |
ADCY9 |
adenylate cyclase 9 |
| chr6_+_132129151 | 0.52 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr5_-_141704566 | 0.48 |
ENST00000344120.4 ENST00000434127.2 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
| chr3_-_129407535 | 0.44 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr6_+_89790490 | 0.43 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr12_-_8088871 | 0.43 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr10_-_735553 | 0.42 |
ENST00000280886.6 ENST00000423550.1 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr20_+_30865429 | 0.41 |
ENST00000375712.3 |
KIF3B |
kinesin family member 3B |
| chr6_-_16761678 | 0.40 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr21_-_28338732 | 0.40 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr5_+_82767284 | 0.39 |
ENST00000265077.3 |
VCAN |
versican |
| chr2_-_97535708 | 0.38 |
ENST00000305476.5 |
SEMA4C |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr8_+_61591337 | 0.38 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr10_-_90712520 | 0.37 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr8_+_26435359 | 0.36 |
ENST00000311151.5 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr12_-_46766577 | 0.36 |
ENST00000256689.5 |
SLC38A2 |
solute carrier family 38, member 2 |
| chr20_+_11871371 | 0.36 |
ENST00000254977.3 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr4_+_88896819 | 0.36 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chrX_-_20284958 | 0.34 |
ENST00000379565.3 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_+_32502952 | 0.33 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr8_-_57123815 | 0.33 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr16_+_15068955 | 0.33 |
ENST00000396410.4 ENST00000569715.1 ENST00000450288.2 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
| chr15_+_80696666 | 0.33 |
ENST00000303329.4 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_+_70480963 | 0.32 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr18_+_55102917 | 0.32 |
ENST00000491143.2 |
ONECUT2 |
one cut homeobox 2 |
| chr17_-_56494713 | 0.32 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr8_-_119964434 | 0.30 |
ENST00000297350.4 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
| chr3_+_186501336 | 0.30 |
ENST00000323963.5 ENST00000440191.2 ENST00000356531.5 |
EIF4A2 |
eukaryotic translation initiation factor 4A2 |
| chr8_-_9008206 | 0.30 |
ENST00000310455.3 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
| chr17_+_28705921 | 0.30 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr1_+_39456895 | 0.29 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr5_-_111093406 | 0.28 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr3_+_187930719 | 0.28 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_69614373 | 0.28 |
ENST00000361060.5 ENST00000357308.4 |
GFPT1 |
glutamine--fructose-6-phosphate transaminase 1 |
| chrX_+_9431324 | 0.28 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr20_+_35201857 | 0.27 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr22_-_39239987 | 0.26 |
ENST00000333039.2 |
NPTXR |
neuronal pentraxin receptor |
| chr22_+_21771656 | 0.26 |
ENST00000407464.2 |
HIC2 |
hypermethylated in cancer 2 |
| chr1_+_40627038 | 0.26 |
ENST00000372771.4 |
RLF |
rearranged L-myc fusion |
| chr4_+_55095264 | 0.26 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr20_+_30697298 | 0.24 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr9_+_470288 | 0.24 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr2_-_26101374 | 0.23 |
ENST00000435504.4 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
| chr5_+_71403061 | 0.23 |
ENST00000512974.1 ENST00000296755.7 |
MAP1B |
microtubule-associated protein 1B |
| chr1_+_28696111 | 0.23 |
ENST00000373839.3 |
PHACTR4 |
phosphatase and actin regulator 4 |
| chr10_+_73975742 | 0.23 |
ENST00000299381.4 |
ANAPC16 |
anaphase promoting complex subunit 16 |
| chr16_-_85045131 | 0.23 |
ENST00000313732.4 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
| chr4_+_184020398 | 0.23 |
ENST00000403733.3 ENST00000378925.3 |
WWC2 |
WW and C2 domain containing 2 |
| chr9_-_130742792 | 0.22 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr5_-_108745689 | 0.22 |
ENST00000361189.2 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
| chr10_+_31608054 | 0.22 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr20_-_47804894 | 0.22 |
ENST00000371828.3 ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
| chr8_-_18871159 | 0.22 |
ENST00000327040.8 ENST00000440756.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr3_-_69101413 | 0.21 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chr16_+_2587998 | 0.21 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr13_+_98605902 | 0.21 |
ENST00000460070.1 ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5 |
importin 5 |
| chr16_-_73082274 | 0.21 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr6_+_64345698 | 0.21 |
ENST00000506783.1 ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3 |
PHD finger protein 3 |
| chr5_+_65018017 | 0.21 |
ENST00000380985.5 ENST00000502464.1 |
NLN |
neurolysin (metallopeptidase M3 family) |
| chr11_-_46940074 | 0.20 |
ENST00000378623.1 ENST00000534404.1 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
| chr14_+_55738021 | 0.20 |
ENST00000313833.4 |
FBXO34 |
F-box protein 34 |
| chr5_-_1524015 | 0.20 |
ENST00000283415.3 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
| chr2_-_122407097 | 0.20 |
ENST00000409078.3 |
CLASP1 |
cytoplasmic linker associated protein 1 |
| chr12_-_12419703 | 0.20 |
ENST00000543091.1 ENST00000261349.4 |
LRP6 |
low density lipoprotein receptor-related protein 6 |
| chr14_+_74111578 | 0.20 |
ENST00000554113.1 ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1 |
dynein, axonemal, light chain 1 |
| chr10_+_69644404 | 0.20 |
ENST00000212015.6 |
SIRT1 |
sirtuin 1 |
| chr20_+_5107420 | 0.19 |
ENST00000460006.1 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr3_+_23986748 | 0.19 |
ENST00000312521.4 |
NR1D2 |
nuclear receptor subfamily 1, group D, member 2 |
| chr7_-_156803329 | 0.19 |
ENST00000252971.6 |
MNX1 |
motor neuron and pancreas homeobox 1 |
| chr3_+_171758344 | 0.19 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr6_-_99797522 | 0.19 |
ENST00000389677.5 |
FAXC |
failed axon connections homolog (Drosophila) |
| chr5_+_137801160 | 0.19 |
ENST00000239938.4 |
EGR1 |
early growth response 1 |
| chr10_+_114709999 | 0.18 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_15413035 | 0.18 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr3_-_48700310 | 0.18 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr12_-_498620 | 0.18 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr11_+_108093839 | 0.18 |
ENST00000452508.2 |
ATM |
ataxia telangiectasia mutated |
| chr16_-_87525651 | 0.18 |
ENST00000268616.4 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
| chr15_+_43803143 | 0.18 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr8_+_21915368 | 0.18 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr6_-_79787902 | 0.18 |
ENST00000275034.4 |
PHIP |
pleckstrin homology domain interacting protein |
| chr20_+_32077880 | 0.18 |
ENST00000342704.6 ENST00000375279.2 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr10_+_89622870 | 0.17 |
ENST00000371953.3 |
PTEN |
phosphatase and tensin homolog |
| chr8_+_37594130 | 0.17 |
ENST00000518526.1 ENST00000523887.1 ENST00000276461.5 |
ERLIN2 |
ER lipid raft associated 2 |
| chr5_+_145826867 | 0.17 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr2_+_64751433 | 0.17 |
ENST00000238856.4 ENST00000422803.1 ENST00000238855.7 |
AFTPH |
aftiphilin |
| chr12_+_94542459 | 0.17 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr22_-_39548627 | 0.17 |
ENST00000216133.5 |
CBX7 |
chromobox homolog 7 |
| chr9_+_110045537 | 0.17 |
ENST00000358015.3 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
| chr6_+_15246501 | 0.16 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr1_-_72748417 | 0.16 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chr11_-_18656028 | 0.16 |
ENST00000336349.5 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr11_+_47291193 | 0.16 |
ENST00000428807.1 ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD |
MAP-kinase activating death domain |
| chr15_+_41952591 | 0.16 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr12_+_6898638 | 0.15 |
ENST00000011653.4 |
CD4 |
CD4 molecule |
| chr12_+_104324112 | 0.15 |
ENST00000299767.5 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr9_+_36036430 | 0.15 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr4_+_38665810 | 0.15 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr8_+_86089460 | 0.14 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr3_+_53195136 | 0.14 |
ENST00000394729.2 ENST00000330452.3 |
PRKCD |
protein kinase C, delta |
| chr20_+_60718785 | 0.14 |
ENST00000421564.1 ENST00000450482.1 ENST00000331758.3 |
SS18L1 |
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr17_+_43971643 | 0.14 |
ENST00000344290.5 ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT |
microtubule-associated protein tau |
| chr5_-_78809950 | 0.14 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr3_-_179169330 | 0.14 |
ENST00000232564.3 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr4_-_120550146 | 0.14 |
ENST00000354960.3 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
| chr8_-_71316021 | 0.14 |
ENST00000452400.2 |
NCOA2 |
nuclear receptor coactivator 2 |
| chr6_+_87865262 | 0.14 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr15_+_45879321 | 0.14 |
ENST00000220531.3 ENST00000567461.1 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr17_-_13505219 | 0.13 |
ENST00000284110.1 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr3_-_142166904 | 0.13 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr1_+_24829384 | 0.13 |
ENST00000374395.4 ENST00000436717.2 |
RCAN3 |
RCAN family member 3 |
| chr9_+_91933407 | 0.13 |
ENST00000375807.3 ENST00000339901.4 |
SECISBP2 |
SECIS binding protein 2 |
| chr17_+_73452545 | 0.13 |
ENST00000314256.7 |
KIAA0195 |
KIAA0195 |
| chr2_-_39348137 | 0.13 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr19_-_5978090 | 0.13 |
ENST00000592621.1 ENST00000034275.8 ENST00000591092.1 ENST00000591333.1 ENST00000590623.1 ENST00000439268.2 ENST00000587159.1 |
RANBP3 |
RAN binding protein 3 |
| chr5_+_79331164 | 0.13 |
ENST00000350881.2 |
THBS4 |
thrombospondin 4 |
| chr1_+_32930647 | 0.13 |
ENST00000609129.1 |
ZBTB8B |
zinc finger and BTB domain containing 8B |
| chr12_+_862089 | 0.12 |
ENST00000315939.6 ENST00000537687.1 ENST00000447667.2 |
WNK1 |
WNK lysine deficient protein kinase 1 |
| chr6_+_10556215 | 0.12 |
ENST00000316170.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr14_+_105781048 | 0.12 |
ENST00000458164.2 ENST00000447393.1 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
| chrX_+_146993449 | 0.12 |
ENST00000218200.8 ENST00000370471.3 ENST00000370477.1 |
FMR1 |
fragile X mental retardation 1 |
| chr7_+_12250886 | 0.12 |
ENST00000444443.1 ENST00000396667.3 |
TMEM106B |
transmembrane protein 106B |
| chr17_-_66287257 | 0.12 |
ENST00000327268.4 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr11_+_12695944 | 0.12 |
ENST00000361905.4 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr10_+_52751010 | 0.12 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr5_+_109025067 | 0.12 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chrX_+_46306624 | 0.12 |
ENST00000360017.5 |
KRBOX4 |
KRAB box domain containing 4 |
| chr18_-_12377283 | 0.12 |
ENST00000269143.3 |
AFG3L2 |
AFG3-like AAA ATPase 2 |
| chr2_-_68479614 | 0.12 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr3_+_37903432 | 0.12 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr1_+_182992545 | 0.12 |
ENST00000258341.4 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
| chr17_-_57970074 | 0.12 |
ENST00000346141.6 |
TUBD1 |
tubulin, delta 1 |
| chr7_+_4721885 | 0.12 |
ENST00000328914.4 |
FOXK1 |
forkhead box K1 |
| chr11_+_125034586 | 0.12 |
ENST00000298282.9 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
| chr10_+_104678032 | 0.12 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr3_-_138553594 | 0.12 |
ENST00000477593.1 ENST00000483968.1 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr6_-_154831779 | 0.12 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr1_+_90286562 | 0.12 |
ENST00000525774.1 ENST00000337338.5 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr3_-_114790179 | 0.12 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr13_-_39612176 | 0.12 |
ENST00000352251.3 ENST00000350125.3 |
PROSER1 |
proline and serine rich 1 |
| chr12_+_102271129 | 0.11 |
ENST00000258534.8 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
| chr5_-_168006591 | 0.11 |
ENST00000239231.6 |
PANK3 |
pantothenate kinase 3 |
| chr22_-_50217981 | 0.11 |
ENST00000457780.2 |
BRD1 |
bromodomain containing 1 |
| chr2_-_176032843 | 0.11 |
ENST00000392544.1 ENST00000409499.1 ENST00000426833.3 ENST00000392543.2 ENST00000538946.1 ENST00000487334.2 ENST00000409833.1 ENST00000409635.1 ENST00000264110.2 ENST00000345739.5 |
ATF2 |
activating transcription factor 2 |
| chr1_-_225840747 | 0.11 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr11_-_30038490 | 0.11 |
ENST00000328224.6 |
KCNA4 |
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr2_+_148602058 | 0.11 |
ENST00000241416.7 ENST00000535787.1 ENST00000404590.1 |
ACVR2A |
activin A receptor, type IIA |
| chr3_+_5020801 | 0.11 |
ENST00000256495.3 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
| chr5_-_98262240 | 0.11 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr5_+_52083730 | 0.11 |
ENST00000282588.6 ENST00000274311.2 |
ITGA1 PELO |
integrin, alpha 1 pelota homolog (Drosophila) |
| chr3_-_195270162 | 0.11 |
ENST00000438848.1 ENST00000328432.3 |
PPP1R2 |
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr3_+_154797428 | 0.11 |
ENST00000460393.1 |
MME |
membrane metallo-endopeptidase |
| chr12_-_66524482 | 0.11 |
ENST00000446587.2 ENST00000266604.2 |
LLPH |
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr1_+_114472222 | 0.11 |
ENST00000369558.1 ENST00000369561.4 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr6_-_136610911 | 0.11 |
ENST00000530767.1 ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1 |
BCL2-associated transcription factor 1 |
| chr4_+_1795012 | 0.11 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr2_-_68694390 | 0.11 |
ENST00000377957.3 |
FBXO48 |
F-box protein 48 |
| chr9_-_36400213 | 0.10 |
ENST00000259605.6 ENST00000353739.4 |
RNF38 |
ring finger protein 38 |
| chr7_+_99613195 | 0.10 |
ENST00000324306.6 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr13_-_76056250 | 0.10 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr6_-_99873145 | 0.10 |
ENST00000369239.5 ENST00000438806.1 |
PNISR |
PNN-interacting serine/arginine-rich protein |
| chr12_-_8025442 | 0.10 |
ENST00000340749.5 ENST00000535295.1 ENST00000539234.1 |
SLC2A14 |
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr1_+_97187318 | 0.10 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr5_+_36876833 | 0.10 |
ENST00000282516.8 ENST00000448238.2 |
NIPBL |
Nipped-B homolog (Drosophila) |
| chr5_-_65017921 | 0.10 |
ENST00000381007.4 |
SGTB |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_-_67525473 | 0.10 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr1_+_93544791 | 0.10 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr6_+_36853607 | 0.09 |
ENST00000480824.2 ENST00000355190.3 ENST00000373685.1 |
C6orf89 |
chromosome 6 open reading frame 89 |
| chr13_-_28545276 | 0.09 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chrX_-_134049262 | 0.09 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr16_-_58663720 | 0.09 |
ENST00000564557.1 ENST00000569240.1 ENST00000441024.2 ENST00000569020.1 ENST00000317147.5 |
CNOT1 |
CCR4-NOT transcription complex, subunit 1 |
| chr2_+_228336849 | 0.09 |
ENST00000409979.2 ENST00000310078.8 |
AGFG1 |
ArfGAP with FG repeats 1 |
| chr10_-_98346801 | 0.09 |
ENST00000371142.4 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
| chr15_-_49447835 | 0.09 |
ENST00000388901.5 ENST00000299259.6 |
COPS2 |
COP9 signalosome subunit 2 |
| chr2_-_47798044 | 0.09 |
ENST00000327876.4 |
KCNK12 |
potassium channel, subfamily K, member 12 |
| chr9_-_104145795 | 0.09 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr2_+_86947296 | 0.09 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr17_+_42148097 | 0.09 |
ENST00000269097.4 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
| chr10_-_11653753 | 0.08 |
ENST00000609104.1 |
USP6NL |
USP6 N-terminal like |
| chr19_+_10982189 | 0.08 |
ENST00000327064.4 ENST00000588947.1 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
| chr2_+_64681219 | 0.08 |
ENST00000238875.5 |
LGALSL |
lectin, galactoside-binding-like |
| chr3_-_53080047 | 0.08 |
ENST00000482396.1 ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1 |
Scm-like with four mbt domains 1 |
| chr9_+_129567282 | 0.08 |
ENST00000449886.1 ENST00000373464.4 ENST00000450858.1 |
ZBTB43 |
zinc finger and BTB domain containing 43 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.2 | 0.6 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.2 | 0.5 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.4 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 0.4 | GO:0072144 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.2 | GO:2001245 | negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.2 | GO:0090244 | trachea cartilage morphogenesis(GO:0060535) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.2 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.2 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.2 | GO:1904884 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.4 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.6 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.1 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:2000301 | regulation of intracellular transport of viral material(GO:1901252) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.1 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0034971 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.0 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.2 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.4 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0043260 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |