Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ARID3A
Z-value: 0.86
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | ARID3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | 0.27 | 3.1e-01 | Click! |
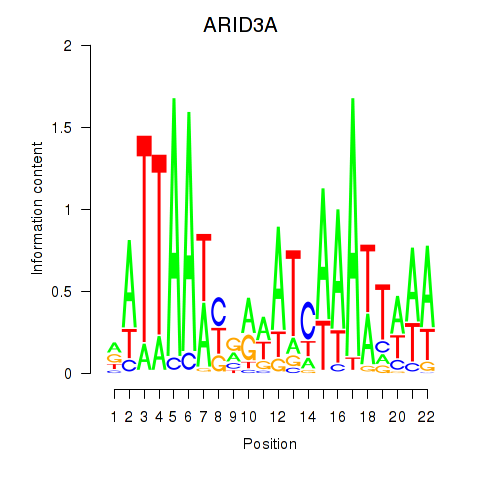
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_165424973 | 1.37 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr3_-_121379739 | 1.10 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr6_+_27791862 | 1.00 |
ENST00000355057.1 |
HIST1H4J |
histone cluster 1, H4j |
| chr4_+_74301880 | 0.96 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr12_+_113344582 | 0.94 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_+_22688150 | 0.83 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr7_+_150264365 | 0.80 |
ENST00000255945.2 ENST00000461940.1 |
GIMAP4 |
GTPase, IMAP family member 4 |
| chr16_+_81812863 | 0.76 |
ENST00000359376.3 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr5_+_121647386 | 0.71 |
ENST00000542191.1 ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr14_+_39734482 | 0.70 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr3_+_43328004 | 0.68 |
ENST00000454177.1 ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK |
SNF related kinase |
| chr17_+_75181292 | 0.65 |
ENST00000431431.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr7_-_139763521 | 0.55 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr2_+_102928009 | 0.54 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr19_-_39826639 | 0.54 |
ENST00000602185.1 ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG |
glia maturation factor, gamma |
| chr10_+_7745232 | 0.49 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745303 | 0.47 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr2_-_160473114 | 0.45 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr7_-_22539771 | 0.43 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr4_+_74606223 | 0.43 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr18_+_61143994 | 0.42 |
ENST00000382771.4 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr17_-_29641104 | 0.41 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr4_-_76957214 | 0.40 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr7_+_115862858 | 0.39 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr9_+_114287433 | 0.38 |
ENST00000358151.4 ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483 |
zinc finger protein 483 |
| chr14_+_24099318 | 0.38 |
ENST00000432832.2 |
DHRS2 |
dehydrogenase/reductase (SDR family) member 2 |
| chr1_+_77333117 | 0.37 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr9_-_69229650 | 0.36 |
ENST00000416428.1 |
CBWD6 |
COBW domain containing 6 |
| chr16_-_21289627 | 0.36 |
ENST00000396023.2 ENST00000415987.2 |
CRYM |
crystallin, mu |
| chr15_-_34880646 | 0.36 |
ENST00000543376.1 |
GOLGA8A |
golgin A8 family, member A |
| chr1_+_207943667 | 0.36 |
ENST00000462968.2 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr4_-_100356291 | 0.36 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_28125638 | 0.35 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr10_-_11574274 | 0.35 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr10_+_62538089 | 0.35 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr4_-_110723194 | 0.34 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr3_+_173116225 | 0.33 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr19_-_48614033 | 0.33 |
ENST00000354276.3 |
PLA2G4C |
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr4_-_110723335 | 0.33 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr17_-_56494713 | 0.33 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr12_-_53601000 | 0.32 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr17_-_56494908 | 0.32 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr8_+_86099884 | 0.31 |
ENST00000517476.1 ENST00000521429.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr3_+_169629354 | 0.31 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
| chr19_-_48614063 | 0.31 |
ENST00000599921.1 ENST00000599111.1 |
PLA2G4C |
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr17_-_56494882 | 0.31 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chrX_-_2882296 | 0.31 |
ENST00000438544.1 ENST00000381134.3 ENST00000545496.1 |
ARSE |
arylsulfatase E (chondrodysplasia punctata 1) |
| chr7_+_89783689 | 0.31 |
ENST00000297205.2 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
| chrY_+_16634483 | 0.31 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr4_+_69962185 | 0.30 |
ENST00000305231.7 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962212 | 0.30 |
ENST00000508661.1 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_-_53387386 | 0.29 |
ENST00000467988.1 ENST00000358358.5 ENST00000371522.4 |
ECHDC2 |
enoyl CoA hydratase domain containing 2 |
| chr8_-_102218292 | 0.29 |
ENST00000518336.1 ENST00000520454.1 |
ZNF706 |
zinc finger protein 706 |
| chr7_+_156902674 | 0.28 |
ENST00000594086.1 |
AC006967.1 |
Protein LOC100996426 |
| chr11_+_67351019 | 0.27 |
ENST00000398606.3 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr11_-_104035088 | 0.27 |
ENST00000302251.5 |
PDGFD |
platelet derived growth factor D |
| chr3_+_118892362 | 0.27 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr18_+_29671812 | 0.27 |
ENST00000261593.3 ENST00000578914.1 |
RNF138 |
ring finger protein 138, E3 ubiquitin protein ligase |
| chr12_-_53601055 | 0.26 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr17_-_29641084 | 0.26 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr1_-_154600421 | 0.26 |
ENST00000368471.3 ENST00000292205.5 |
ADAR |
adenosine deaminase, RNA-specific |
| chr5_+_59783941 | 0.25 |
ENST00000506884.1 ENST00000504876.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_241695670 | 0.25 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_90198535 | 0.24 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr16_-_21868978 | 0.24 |
ENST00000357370.5 ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr16_-_29415350 | 0.24 |
ENST00000524087.1 |
NPIPB11 |
nuclear pore complex interacting protein family, member B11 |
| chr1_-_207206092 | 0.23 |
ENST00000359470.5 ENST00000461135.2 |
C1orf116 |
chromosome 1 open reading frame 116 |
| chr12_-_81763184 | 0.23 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr12_-_81763127 | 0.23 |
ENST00000541017.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_-_106725723 | 0.23 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr7_+_107224364 | 0.23 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr14_-_53162361 | 0.22 |
ENST00000395686.3 |
ERO1L |
ERO1-like (S. cerevisiae) |
| chr15_+_75491213 | 0.22 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr6_+_31105426 | 0.22 |
ENST00000547221.1 |
PSORS1C1 |
psoriasis susceptibility 1 candidate 1 |
| chr8_-_2585929 | 0.22 |
ENST00000519393.1 ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1 |
RP11-134O21.1 |
| chr9_-_125590818 | 0.21 |
ENST00000259467.4 |
PDCL |
phosducin-like |
| chr9_-_70465758 | 0.21 |
ENST00000489273.1 |
CBWD5 |
COBW domain containing 5 |
| chr11_+_71249071 | 0.20 |
ENST00000398534.3 |
KRTAP5-8 |
keratin associated protein 5-8 |
| chr1_+_158900568 | 0.20 |
ENST00000458222.1 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr12_+_54892550 | 0.20 |
ENST00000545638.2 |
NCKAP1L |
NCK-associated protein 1-like |
| chr4_+_100737954 | 0.19 |
ENST00000296414.7 ENST00000512369.1 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr15_-_45670924 | 0.19 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_+_26150628 | 0.19 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr10_+_26727125 | 0.19 |
ENST00000376236.4 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr2_+_110551851 | 0.19 |
ENST00000272454.6 ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5 |
RANBP2-like and GRIP domain containing 5 |
| chr5_-_107703556 | 0.19 |
ENST00000496714.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr17_-_18430160 | 0.18 |
ENST00000392176.3 |
FAM106A |
family with sequence similarity 106, member A |
| chr2_-_89340242 | 0.18 |
ENST00000480492.1 |
IGKV1-12 |
immunoglobulin kappa variable 1-12 |
| chr1_+_12976450 | 0.18 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr2_+_109335929 | 0.18 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
| chr5_+_54455946 | 0.18 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr14_-_50101931 | 0.17 |
ENST00000298292.8 ENST00000406043.3 |
DNAAF2 |
dynein, axonemal, assembly factor 2 |
| chr9_+_42671887 | 0.17 |
ENST00000456520.1 ENST00000377391.3 |
CBWD7 |
COBW domain containing 7 |
| chr2_+_106468204 | 0.17 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr16_-_21868739 | 0.17 |
ENST00000415645.2 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr10_-_69597915 | 0.17 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr15_+_25068773 | 0.17 |
ENST00000400100.1 ENST00000400098.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr12_-_68696652 | 0.17 |
ENST00000539972.1 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr1_+_53308398 | 0.16 |
ENST00000371528.1 |
ZYG11A |
zyg-11 family member A, cell cycle regulator |
| chr2_-_191115229 | 0.16 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chr9_+_90112117 | 0.16 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr20_-_33732952 | 0.16 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_+_32832134 | 0.16 |
ENST00000452533.2 |
DNM1L |
dynamin 1-like |
| chr2_-_208994548 | 0.16 |
ENST00000282141.3 |
CRYGC |
crystallin, gamma C |
| chr2_+_210444142 | 0.16 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr6_+_24403144 | 0.16 |
ENST00000274747.7 ENST00000543597.1 ENST00000535061.1 ENST00000378353.1 ENST00000378386.3 ENST00000443868.2 |
MRS2 |
MRS2 magnesium transporter |
| chr6_-_30181133 | 0.15 |
ENST00000454678.2 ENST00000434785.1 |
TRIM26 |
tripartite motif containing 26 |
| chr2_-_111334678 | 0.15 |
ENST00000329516.3 ENST00000330331.5 ENST00000446930.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chrX_-_64254587 | 0.15 |
ENST00000337990.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr12_+_26126681 | 0.15 |
ENST00000542865.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_+_33439268 | 0.15 |
ENST00000594612.1 |
FKSG48 |
FKSG48 |
| chr13_-_31040060 | 0.15 |
ENST00000326004.4 ENST00000341423.5 |
HMGB1 |
high mobility group box 1 |
| chr8_+_104892639 | 0.15 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr1_+_73771844 | 0.15 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr10_-_100995540 | 0.15 |
ENST00000370546.1 ENST00000404542.1 |
HPSE2 |
heparanase 2 |
| chr13_-_31039375 | 0.15 |
ENST00000399494.1 |
HMGB1 |
high mobility group box 1 |
| chr2_-_183106641 | 0.15 |
ENST00000346717.4 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_15466850 | 0.15 |
ENST00000438826.3 ENST00000225576.3 ENST00000519970.1 ENST00000518321.1 ENST00000428082.2 ENST00000522212.2 |
TVP23C TVP23C-CDRT4 |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) TVP23C-CDRT4 readthrough |
| chr16_-_85722530 | 0.15 |
ENST00000253462.3 |
GINS2 |
GINS complex subunit 2 (Psf2 homolog) |
| chr6_-_46703430 | 0.14 |
ENST00000537365.1 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_44588877 | 0.14 |
ENST00000576629.1 |
LRRC37A2 |
leucine rich repeat containing 37, member A2 |
| chr3_+_46395219 | 0.14 |
ENST00000445132.2 ENST00000292301.4 |
CCR2 |
chemokine (C-C motif) receptor 2 |
| chr6_-_64029879 | 0.14 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr19_+_45542295 | 0.14 |
ENST00000221455.3 ENST00000391953.4 ENST00000588936.1 |
CLASRP |
CLK4-associating serine/arginine rich protein |
| chr1_-_39347255 | 0.14 |
ENST00000454994.2 ENST00000357771.3 |
GJA9 |
gap junction protein, alpha 9, 59kDa |
| chr16_-_20753114 | 0.14 |
ENST00000396083.2 |
THUMPD1 |
THUMP domain containing 1 |
| chr1_-_157014865 | 0.14 |
ENST00000361409.2 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr6_-_30181156 | 0.14 |
ENST00000418026.1 ENST00000416596.1 ENST00000453195.1 |
TRIM26 |
tripartite motif containing 26 |
| chr21_-_37270727 | 0.14 |
ENST00000599809.1 |
FKSG68 |
FKSG68 |
| chr19_-_13617247 | 0.13 |
ENST00000573710.2 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_-_157015162 | 0.13 |
ENST00000368194.3 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_+_138340049 | 0.13 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr13_-_79233314 | 0.13 |
ENST00000282003.6 |
RNF219 |
ring finger protein 219 |
| chr8_-_16035454 | 0.13 |
ENST00000355282.2 |
MSR1 |
macrophage scavenger receptor 1 |
| chr1_-_151162606 | 0.12 |
ENST00000354473.4 ENST00000368892.4 |
VPS72 |
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr2_+_210444748 | 0.12 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr15_-_38519066 | 0.12 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr5_+_159656437 | 0.12 |
ENST00000402432.3 |
FABP6 |
fatty acid binding protein 6, ileal |
| chr6_+_42749759 | 0.12 |
ENST00000314073.5 |
GLTSCR1L |
GLTSCR1-like |
| chrX_+_83116142 | 0.12 |
ENST00000329312.4 |
CYLC1 |
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr7_-_124569991 | 0.12 |
ENST00000446993.1 ENST00000357628.3 ENST00000393329.1 |
POT1 |
protection of telomeres 1 |
| chr10_-_100995603 | 0.12 |
ENST00000370552.3 ENST00000370549.1 |
HPSE2 |
heparanase 2 |
| chr12_+_53399942 | 0.11 |
ENST00000262056.9 |
EIF4B |
eukaryotic translation initiation factor 4B |
| chr1_-_180991978 | 0.11 |
ENST00000542060.1 ENST00000258301.5 |
STX6 |
syntaxin 6 |
| chr10_-_115904361 | 0.11 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr20_+_57226284 | 0.11 |
ENST00000458280.1 ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16 |
syntaxin 16 |
| chr20_-_60573188 | 0.11 |
ENST00000474089.1 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chrX_-_80457385 | 0.11 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr10_-_48806939 | 0.11 |
ENST00000374233.3 ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B |
protein tyrosine phosphatase, non-receptor type 20B |
| chr2_-_89399845 | 0.11 |
ENST00000479981.1 |
IGKV1-16 |
immunoglobulin kappa variable 1-16 |
| chrX_+_41192595 | 0.11 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr19_+_11200038 | 0.11 |
ENST00000558518.1 ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR |
low density lipoprotein receptor |
| chr12_+_32832203 | 0.11 |
ENST00000553257.1 ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L |
dynamin 1-like |
| chr10_+_51565108 | 0.11 |
ENST00000438493.1 ENST00000452682.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr12_+_133757995 | 0.11 |
ENST00000536435.2 ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268 |
zinc finger protein 268 |
| chr6_-_136847099 | 0.11 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr13_-_88323218 | 0.11 |
ENST00000436290.2 ENST00000453832.2 ENST00000606590.1 |
MIR4500HG |
MIR4500 host gene (non-protein coding) |
| chr16_-_21436459 | 0.11 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr14_+_97263641 | 0.11 |
ENST00000216639.3 |
VRK1 |
vaccinia related kinase 1 |
| chr1_-_100231349 | 0.11 |
ENST00000287474.5 ENST00000414213.1 |
FRRS1 |
ferric-chelate reductase 1 |
| chr10_-_69597810 | 0.10 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr13_+_98612446 | 0.10 |
ENST00000496368.1 ENST00000421861.2 ENST00000357602.3 |
IPO5 |
importin 5 |
| chr12_-_102591604 | 0.10 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr19_-_53758094 | 0.10 |
ENST00000601828.1 ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677 |
zinc finger protein 677 |
| chr17_-_15466742 | 0.10 |
ENST00000584811.1 ENST00000419890.2 |
TVP23C |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr10_+_51565188 | 0.10 |
ENST00000430396.2 ENST00000374087.4 ENST00000414907.2 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr12_+_10658201 | 0.09 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chrX_-_15333736 | 0.09 |
ENST00000380470.3 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr6_-_32977345 | 0.09 |
ENST00000450833.2 ENST00000374813.1 ENST00000229829.5 |
HLA-DOA |
major histocompatibility complex, class II, DO alpha |
| chr18_+_11857439 | 0.09 |
ENST00000602628.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr14_-_69261310 | 0.09 |
ENST00000336440.3 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr16_-_11375179 | 0.09 |
ENST00000312511.3 |
PRM1 |
protamine 1 |
| chr9_-_19786926 | 0.09 |
ENST00000341998.2 ENST00000286344.3 |
SLC24A2 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr9_+_21440440 | 0.08 |
ENST00000276927.1 |
IFNA1 |
interferon, alpha 1 |
| chr4_-_185275104 | 0.08 |
ENST00000317596.3 |
RP11-290F5.2 |
RP11-290F5.2 |
| chr10_-_46620012 | 0.08 |
ENST00000508602.1 ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A |
protein tyrosine phosphatase, non-receptor type 20A |
| chr9_-_86571628 | 0.08 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr5_+_140180635 | 0.08 |
ENST00000522353.2 ENST00000532566.2 |
PCDHA3 |
protocadherin alpha 3 |
| chr19_+_36142147 | 0.08 |
ENST00000590618.1 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr19_+_18208603 | 0.08 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr5_-_10308125 | 0.08 |
ENST00000296658.3 |
CMBL |
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chrX_+_49091920 | 0.08 |
ENST00000376227.3 |
CCDC22 |
coiled-coil domain containing 22 |
| chr15_-_37390482 | 0.08 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr17_+_41006095 | 0.08 |
ENST00000591562.1 ENST00000588033.1 |
AOC3 |
amine oxidase, copper containing 3 |
| chr5_-_59783882 | 0.08 |
ENST00000505507.2 ENST00000502484.2 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr6_+_118869452 | 0.07 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr1_-_92371839 | 0.07 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr4_+_71296204 | 0.07 |
ENST00000413702.1 |
MUC7 |
mucin 7, secreted |
| chr7_-_104909435 | 0.07 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr12_-_7848364 | 0.07 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr5_-_16916624 | 0.07 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr18_+_616672 | 0.07 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr7_+_138943265 | 0.07 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chrX_-_63005405 | 0.07 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr1_+_224544572 | 0.06 |
ENST00000366857.5 ENST00000366856.3 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
| chr19_+_15852203 | 0.06 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr2_+_119699864 | 0.06 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr6_+_46761118 | 0.06 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr9_-_139343294 | 0.06 |
ENST00000313084.5 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chr7_+_16828866 | 0.06 |
ENST00000597084.1 |
AC073333.1 |
Uncharacterized protein |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.4 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.2 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 0.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 1.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0002316 | follicular B cell differentiation(GO:0002316) activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 1.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.7 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.6 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.3 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 1.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.2 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0033591 | response to L-ascorbic acid(GO:0033591) negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.4 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.2 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.1 | GO:0051096 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.5 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.0 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |