Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ATF5
Z-value: 0.72
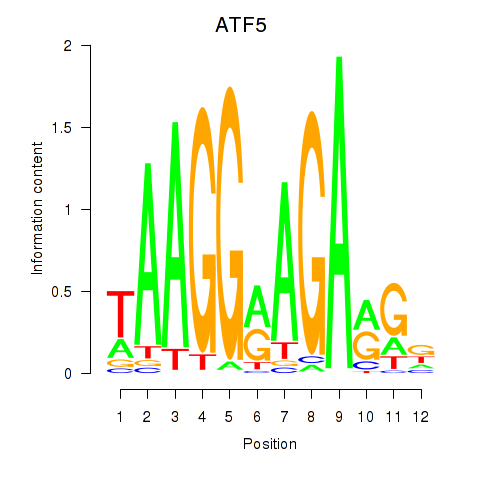
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | ATF5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50432400_50432479 | -0.43 | 9.5e-02 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_65155979 | 2.35 |
ENST00000562325.1 ENST00000268603.4 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr7_-_41740181 | 2.13 |
ENST00000442711.1 |
INHBA |
inhibin, beta A |
| chr2_-_190927447 | 1.83 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr16_-_65155833 | 1.69 |
ENST00000566827.1 ENST00000394156.3 ENST00000562998.1 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr2_+_189839046 | 1.45 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chrX_-_13835147 | 1.39 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr2_+_217498105 | 1.37 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr1_-_103574024 | 1.31 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr22_-_36236623 | 1.31 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr22_-_36236265 | 1.19 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr5_+_34757309 | 1.14 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr3_-_99569821 | 1.09 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr15_+_80733570 | 1.07 |
ENST00000533983.1 ENST00000527771.1 ENST00000525103.1 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr3_-_127441406 | 0.96 |
ENST00000487473.1 ENST00000484451.1 |
MGLL |
monoglyceride lipase |
| chr1_-_17304771 | 0.89 |
ENST00000375534.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr5_+_82767487 | 0.85 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_+_82767284 | 0.85 |
ENST00000265077.3 |
VCAN |
versican |
| chr12_-_52967600 | 0.85 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr8_+_79428539 | 0.76 |
ENST00000352966.5 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr10_-_75415825 | 0.73 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr8_-_13134045 | 0.66 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr5_+_82767583 | 0.65 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr8_+_22462532 | 0.61 |
ENST00000389279.3 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
| chr9_-_23825956 | 0.60 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr7_+_128399002 | 0.56 |
ENST00000493278.1 |
CALU |
calumenin |
| chr8_+_22462145 | 0.55 |
ENST00000308511.4 ENST00000523801.1 ENST00000521301.1 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
| chr3_+_101504200 | 0.53 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr22_-_36357671 | 0.53 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chrX_-_13835461 | 0.53 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr7_-_37956409 | 0.52 |
ENST00000436072.2 |
SFRP4 |
secreted frizzled-related protein 4 |
| chr11_+_124735282 | 0.50 |
ENST00000397801.1 |
ROBO3 |
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr13_-_36944307 | 0.46 |
ENST00000355182.4 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr12_+_56915776 | 0.46 |
ENST00000550726.1 ENST00000542360.1 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr18_-_53089723 | 0.43 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr16_+_19567016 | 0.42 |
ENST00000251143.5 ENST00000417362.2 ENST00000567245.1 ENST00000513947.4 |
C16orf62 |
chromosome 16 open reading frame 62 |
| chr12_+_56915713 | 0.41 |
ENST00000262031.5 ENST00000552247.2 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
| chr5_+_137203465 | 0.38 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr12_+_104680659 | 0.38 |
ENST00000526691.1 ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1 |
thioredoxin reductase 1 |
| chr19_-_45926739 | 0.36 |
ENST00000589381.1 ENST00000591636.1 ENST00000013807.5 ENST00000592023.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr2_-_145278475 | 0.35 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr9_-_23826298 | 0.35 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_47022140 | 0.34 |
ENST00000290330.3 |
SNF8 |
SNF8, ESCRT-II complex subunit |
| chr10_+_135207598 | 0.32 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr14_+_96968802 | 0.31 |
ENST00000556619.1 ENST00000392990.2 |
PAPOLA |
poly(A) polymerase alpha |
| chr8_+_95732095 | 0.31 |
ENST00000414645.2 |
DPY19L4 |
dpy-19-like 4 (C. elegans) |
| chr6_+_114178512 | 0.29 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr14_+_96968707 | 0.29 |
ENST00000216277.8 ENST00000557320.1 ENST00000557471.1 |
PAPOLA |
poly(A) polymerase alpha |
| chr2_-_188419200 | 0.29 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_+_88047606 | 0.28 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr3_-_69129501 | 0.27 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
| chr2_-_87018784 | 0.27 |
ENST00000283635.3 ENST00000538832.1 |
CD8A |
CD8a molecule |
| chr11_+_27076764 | 0.26 |
ENST00000525090.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_-_65569244 | 0.26 |
ENST00000557277.1 ENST00000556892.1 |
MAX |
MYC associated factor X |
| chr10_+_111765562 | 0.25 |
ENST00000360162.3 |
ADD3 |
adducin 3 (gamma) |
| chr10_+_135207623 | 0.25 |
ENST00000317502.6 ENST00000432508.3 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr1_-_27930102 | 0.24 |
ENST00000247087.5 ENST00000374011.2 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
| chr2_-_87248975 | 0.24 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr12_-_72057638 | 0.22 |
ENST00000552037.1 ENST00000378743.3 |
ZFC3H1 |
zinc finger, C3H1-type containing |
| chr18_-_812517 | 0.22 |
ENST00000584307.1 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr3_-_88108212 | 0.22 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr1_+_36023035 | 0.21 |
ENST00000373253.3 |
NCDN |
neurochondrin |
| chr6_-_100016492 | 0.20 |
ENST00000369217.4 ENST00000369220.4 ENST00000482541.2 |
CCNC |
cyclin C |
| chr6_-_31514333 | 0.20 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr3_-_47950745 | 0.20 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr14_-_65569057 | 0.19 |
ENST00000555419.1 ENST00000341653.2 |
MAX |
MYC associated factor X |
| chr17_+_55055466 | 0.19 |
ENST00000262288.3 ENST00000572710.1 ENST00000575395.1 |
SCPEP1 |
serine carboxypeptidase 1 |
| chr2_-_197036289 | 0.19 |
ENST00000263955.4 |
STK17B |
serine/threonine kinase 17b |
| chr3_+_38206975 | 0.18 |
ENST00000446845.1 ENST00000311806.3 |
OXSR1 |
oxidative stress responsive 1 |
| chr1_+_36023370 | 0.18 |
ENST00000356090.4 ENST00000373243.2 |
NCDN |
neurochondrin |
| chr2_-_113542063 | 0.17 |
ENST00000263339.3 |
IL1A |
interleukin 1, alpha |
| chr2_-_100759037 | 0.17 |
ENST00000317233.4 ENST00000423966.1 ENST00000416492.1 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr18_-_812231 | 0.17 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr12_-_16761007 | 0.16 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr14_+_79745746 | 0.15 |
ENST00000281127.7 |
NRXN3 |
neurexin 3 |
| chr2_-_26700900 | 0.15 |
ENST00000338581.6 ENST00000339598.3 ENST00000402415.3 |
OTOF |
otoferlin |
| chr12_-_12491608 | 0.14 |
ENST00000545735.1 |
MANSC1 |
MANSC domain containing 1 |
| chr3_-_88108192 | 0.14 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr14_+_70918874 | 0.14 |
ENST00000603540.1 |
ADAM21 |
ADAM metallopeptidase domain 21 |
| chr16_-_30023615 | 0.14 |
ENST00000564979.1 ENST00000563378.1 |
DOC2A |
double C2-like domains, alpha |
| chr17_+_33914460 | 0.14 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_26205340 | 0.13 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr3_+_23958470 | 0.13 |
ENST00000434031.2 ENST00000413699.1 ENST00000456530.2 |
RPL15 |
ribosomal protein L15 |
| chr6_-_100016678 | 0.12 |
ENST00000523799.1 ENST00000520429.1 |
CCNC |
cyclin C |
| chr14_-_65569186 | 0.12 |
ENST00000555932.1 ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX |
MYC associated factor X |
| chr8_-_131028869 | 0.12 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr10_+_103113840 | 0.12 |
ENST00000393441.4 ENST00000408038.2 |
BTRC |
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr1_-_183387723 | 0.11 |
ENST00000287713.6 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
| chr17_+_38334242 | 0.11 |
ENST00000436615.3 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr17_+_33914276 | 0.10 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr6_-_39902160 | 0.10 |
ENST00000340692.5 |
MOCS1 |
molybdenum cofactor synthesis 1 |
| chr1_-_46598371 | 0.10 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr19_-_45873642 | 0.09 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr5_+_138629337 | 0.08 |
ENST00000394805.3 ENST00000512876.1 ENST00000513678.1 |
MATR3 |
matrin 3 |
| chr5_-_132200477 | 0.08 |
ENST00000296875.2 |
GDF9 |
growth differentiation factor 9 |
| chr20_+_54987168 | 0.07 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr6_-_100016527 | 0.07 |
ENST00000523985.1 ENST00000518714.1 ENST00000520371.1 |
CCNC |
cyclin C |
| chr1_+_151739131 | 0.07 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chrX_+_153409678 | 0.07 |
ENST00000369951.4 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
| chr3_-_183145765 | 0.06 |
ENST00000473233.1 |
MCF2L2 |
MCF.2 cell line derived transforming sequence-like 2 |
| chr5_+_138629389 | 0.06 |
ENST00000504045.1 ENST00000504311.1 ENST00000502499.1 |
MATR3 |
matrin 3 |
| chr7_-_115670804 | 0.05 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr15_+_63481668 | 0.05 |
ENST00000321437.4 ENST00000559006.1 ENST00000448330.2 |
RAB8B |
RAB8B, member RAS oncogene family |
| chr12_-_80084594 | 0.05 |
ENST00000548426.1 |
PAWR |
PRKC, apoptosis, WT1, regulator |
| chr3_+_19988885 | 0.05 |
ENST00000422242.1 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr11_-_106889250 | 0.05 |
ENST00000526355.2 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
| chr1_+_202091980 | 0.05 |
ENST00000367282.5 |
GPR37L1 |
G protein-coupled receptor 37 like 1 |
| chr11_-_106889157 | 0.05 |
ENST00000282249.2 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
| chr12_-_25101920 | 0.04 |
ENST00000539780.1 ENST00000546285.1 ENST00000342945.5 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr15_-_90222610 | 0.04 |
ENST00000300055.5 |
PLIN1 |
perilipin 1 |
| chr8_+_100025476 | 0.04 |
ENST00000355155.1 ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
| chr20_+_54987305 | 0.04 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr20_+_43211149 | 0.04 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr15_-_90222642 | 0.03 |
ENST00000430628.2 |
PLIN1 |
perilipin 1 |
| chr4_+_77172847 | 0.03 |
ENST00000515604.1 ENST00000539752.1 ENST00000424749.2 |
FAM47E FAM47E-STBD1 FAM47E |
uncharacterized protein LOC100631383 FAM47E-STBD1 readthrough family with sequence similarity 47, member E |
| chr4_+_71108300 | 0.03 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr7_-_115670792 | 0.03 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr3_-_183145873 | 0.03 |
ENST00000447025.2 ENST00000414362.2 ENST00000328913.3 |
MCF2L2 |
MCF.2 cell line derived transforming sequence-like 2 |
| chrX_+_153448107 | 0.03 |
ENST00000369935.5 |
OPN1MW |
opsin 1 (cone pigments), medium-wave-sensitive |
| chr12_-_120884175 | 0.03 |
ENST00000546954.1 |
TRIAP1 |
TP53 regulated inhibitor of apoptosis 1 |
| chrX_-_47489244 | 0.02 |
ENST00000469388.1 ENST00000396992.3 ENST00000377005.2 |
CFP |
complement factor properdin |
| chr12_-_49351303 | 0.02 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chr17_+_7533439 | 0.02 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr3_-_62861012 | 0.02 |
ENST00000357948.3 ENST00000383710.4 |
CADPS |
Ca++-dependent secretion activator |
| chr15_+_77287426 | 0.02 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr1_+_158323755 | 0.02 |
ENST00000368157.1 ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E |
CD1e molecule |
| chr5_+_137203557 | 0.01 |
ENST00000515645.1 |
MYOT |
myotilin |
| chr5_+_137203541 | 0.01 |
ENST00000421631.2 |
MYOT |
myotilin |
| chr17_-_46691990 | 0.01 |
ENST00000576562.1 |
HOXB8 |
homeobox B8 |
| chr17_+_38219063 | 0.01 |
ENST00000584985.1 ENST00000264637.4 ENST00000450525.2 |
THRA |
thyroid hormone receptor, alpha |
| chr4_-_175750364 | 0.01 |
ENST00000340217.5 ENST00000274093.3 |
GLRA3 |
glycine receptor, alpha 3 |
| chr19_+_10531150 | 0.00 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr5_+_175223313 | 0.00 |
ENST00000359546.4 |
CPLX2 |
complexin 2 |
| chr3_+_19988566 | 0.00 |
ENST00000273047.4 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr5_+_92919043 | 0.00 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 2.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 2.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0070522 | nucleotide-excision repair factor 1 complex(GO:0000110) ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 3.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 1.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.4 | 4.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 1.9 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 1.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 1.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.5 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 1.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 2.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.3 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.2 | GO:1901978 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 0.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.2 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.7 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.4 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:2000391 | ectopic germ cell programmed cell death(GO:0035234) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0030828 | positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.1 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.4 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.5 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0051036 | regulation of endosome size(GO:0051036) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 3.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |