Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
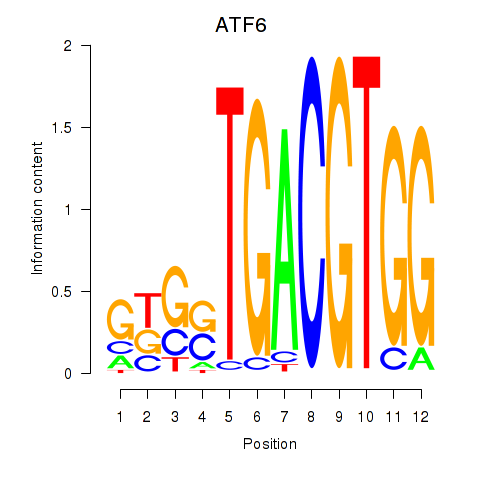
Results for ATF6
Z-value: 1.14
Transcription factors associated with ATF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF6
|
ENSG00000118217.5 | ATF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF6 | hg19_v2_chr1_+_161736072_161736093 | 0.09 | 7.5e-01 | Click! |
Activity profile of ATF6 motif
Sorted Z-values of ATF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_100914781 | 5.01 |
ENST00000431597.1 ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2 |
armadillo repeat containing, X-linked 2 |
| chr6_+_116692102 | 3.60 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr11_-_35547572 | 2.59 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr11_-_35547151 | 2.34 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr12_-_106641728 | 2.28 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chrX_+_102631844 | 2.26 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chrX_+_102631248 | 2.16 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr14_+_96505659 | 2.05 |
ENST00000555004.1 |
C14orf132 |
chromosome 14 open reading frame 132 |
| chr19_+_16187085 | 1.90 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr9_+_112542572 | 1.89 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr9_+_36036430 | 1.78 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr19_+_16186903 | 1.73 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chr8_+_94929077 | 1.54 |
ENST00000297598.4 ENST00000520614.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr9_-_79520989 | 1.52 |
ENST00000376713.3 ENST00000376718.3 ENST00000428286.1 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr8_+_94929168 | 1.52 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_+_94929110 | 1.47 |
ENST00000520728.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr7_-_19157248 | 1.42 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr11_+_32112431 | 1.40 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr21_+_38445539 | 1.38 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr22_+_38864041 | 1.31 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr13_-_49107303 | 1.31 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr8_-_98290087 | 1.31 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr19_+_49458107 | 1.28 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr20_-_34042558 | 1.22 |
ENST00000374372.1 |
GDF5 |
growth differentiation factor 5 |
| chr15_+_67420441 | 1.20 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr5_-_72744336 | 1.18 |
ENST00000499003.3 |
FOXD1 |
forkhead box D1 |
| chrX_-_100872911 | 1.17 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr3_+_45067659 | 1.11 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr7_-_86849883 | 1.07 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr16_-_66959429 | 0.98 |
ENST00000420652.1 ENST00000299759.6 |
RRAD |
Ras-related associated with diabetes |
| chr5_-_131562935 | 0.98 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_+_33318914 | 0.97 |
ENST00000484457.1 ENST00000538892.1 ENST00000538181.1 ENST00000446237.3 ENST00000507198.1 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
| chr4_-_119757239 | 0.97 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr4_-_111544254 | 0.97 |
ENST00000306732.3 |
PITX2 |
paired-like homeodomain 2 |
| chr10_-_118032979 | 0.97 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr1_-_94050668 | 0.97 |
ENST00000539242.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr18_+_56530794 | 0.92 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr3_+_50284321 | 0.90 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr3_+_100211412 | 0.89 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chr1_+_19970657 | 0.84 |
ENST00000375136.3 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
| chr1_-_152009460 | 0.84 |
ENST00000271638.2 |
S100A11 |
S100 calcium binding protein A11 |
| chr7_+_128470431 | 0.83 |
ENST00000325888.8 ENST00000346177.6 |
FLNC |
filamin C, gamma |
| chr4_+_668348 | 0.83 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr7_+_100464760 | 0.81 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr6_-_128841503 | 0.81 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr1_+_87170577 | 0.79 |
ENST00000482504.1 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
| chr1_+_19970797 | 0.78 |
ENST00000548815.1 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
| chr17_-_39968406 | 0.76 |
ENST00000393928.1 |
LEPREL4 |
leprecan-like 4 |
| chr19_-_10121144 | 0.76 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr14_+_64970662 | 0.76 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr9_-_89562104 | 0.76 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr5_+_34656569 | 0.75 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr5_+_34656331 | 0.73 |
ENST00000265109.3 |
RAI14 |
retinoic acid induced 14 |
| chr12_-_102455902 | 0.73 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr11_+_1891380 | 0.73 |
ENST00000429923.1 ENST00000418975.1 ENST00000406638.2 |
LSP1 |
lymphocyte-specific protein 1 |
| chr12_-_102455846 | 0.73 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr15_-_40213080 | 0.72 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr7_+_128379346 | 0.71 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr1_-_20812690 | 0.70 |
ENST00000375078.3 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr8_-_57906362 | 0.70 |
ENST00000262644.4 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
| chr14_+_50087468 | 0.70 |
ENST00000305386.2 |
MGAT2 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr13_-_36920615 | 0.70 |
ENST00000494062.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr2_+_242254679 | 0.69 |
ENST00000428282.1 ENST00000360051.3 |
SEPT2 |
septin 2 |
| chrX_+_48433326 | 0.69 |
ENST00000376755.1 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr3_-_57583185 | 0.67 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr13_-_36920872 | 0.65 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr13_-_36920420 | 0.64 |
ENST00000438666.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr17_-_66951474 | 0.64 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr3_+_129158926 | 0.64 |
ENST00000347300.2 ENST00000296266.3 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr7_-_37956409 | 0.64 |
ENST00000436072.2 |
SFRP4 |
secreted frizzled-related protein 4 |
| chr2_-_242255117 | 0.60 |
ENST00000420451.1 ENST00000417540.1 ENST00000310931.4 |
HDLBP |
high density lipoprotein binding protein |
| chr7_+_128379449 | 0.60 |
ENST00000479257.1 |
CALU |
calumenin |
| chr10_+_112257596 | 0.59 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr3_-_57583130 | 0.58 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr12_-_56123444 | 0.58 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr3_-_121468513 | 0.57 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr3_-_57583052 | 0.57 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_+_148709128 | 0.57 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr16_-_88923285 | 0.57 |
ENST00000542788.1 ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr5_-_131563501 | 0.56 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_+_158519654 | 0.56 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr1_-_161993616 | 0.56 |
ENST00000294794.3 |
OLFML2B |
olfactomedin-like 2B |
| chr2_-_55276320 | 0.55 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr3_+_133292574 | 0.54 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr2_-_98612350 | 0.54 |
ENST00000186436.5 |
TMEM131 |
transmembrane protein 131 |
| chr9_+_114393634 | 0.54 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr3_+_129159039 | 0.54 |
ENST00000507564.1 ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122 |
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr3_-_121468602 | 0.53 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr2_+_242254507 | 0.52 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chr2_-_197036289 | 0.51 |
ENST00000263955.4 |
STK17B |
serine/threonine kinase 17b |
| chr17_-_39968855 | 0.51 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr2_-_237076992 | 0.50 |
ENST00000306318.4 |
GBX2 |
gastrulation brain homeobox 2 |
| chr17_-_19771216 | 0.50 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr14_-_23822080 | 0.48 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr6_-_90062543 | 0.48 |
ENST00000435041.2 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
| chr21_-_38445470 | 0.47 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr13_+_114238997 | 0.47 |
ENST00000538138.1 ENST00000375370.5 |
TFDP1 |
transcription factor Dp-1 |
| chr10_-_74856608 | 0.46 |
ENST00000307116.2 ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1 |
prolyl 4-hydroxylase, alpha polypeptide I |
| chr19_-_48018203 | 0.44 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr14_-_23822061 | 0.44 |
ENST00000397260.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr17_+_33914276 | 0.44 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr17_-_57184260 | 0.43 |
ENST00000376149.3 ENST00000393066.3 |
TRIM37 |
tripartite motif containing 37 |
| chr17_+_33914460 | 0.43 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr17_-_17875688 | 0.42 |
ENST00000379504.3 ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2 |
target of myb1-like 2 (chicken) |
| chr2_+_242255297 | 0.42 |
ENST00000401990.1 ENST00000407971.1 ENST00000436795.1 ENST00000411484.1 ENST00000434955.1 ENST00000402092.2 ENST00000441533.1 ENST00000443492.1 ENST00000437066.1 ENST00000429791.1 |
SEPT2 |
septin 2 |
| chr6_-_86352982 | 0.42 |
ENST00000369622.3 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr12_-_125398654 | 0.42 |
ENST00000541645.1 ENST00000540351.1 |
UBC |
ubiquitin C |
| chr21_-_38445443 | 0.42 |
ENST00000360525.4 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr15_+_23255242 | 0.41 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr19_+_13858593 | 0.41 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr3_-_156272924 | 0.41 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr6_+_127587755 | 0.40 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr8_+_22423168 | 0.40 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr17_-_57184064 | 0.40 |
ENST00000262294.7 |
TRIM37 |
tripartite motif containing 37 |
| chr12_+_54378923 | 0.39 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr12_+_147052 | 0.39 |
ENST00000594563.1 |
AC026369.1 |
Uncharacterized protein |
| chr15_+_31619013 | 0.39 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr1_-_160313025 | 0.38 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr7_+_99699280 | 0.38 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr17_+_18684563 | 0.37 |
ENST00000476139.1 |
TVP23B |
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr12_-_125398602 | 0.37 |
ENST00000541272.1 ENST00000535131.1 |
UBC |
ubiquitin C |
| chr1_-_32169761 | 0.36 |
ENST00000271069.6 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chrX_+_153672468 | 0.36 |
ENST00000393600.3 |
FAM50A |
family with sequence similarity 50, member A |
| chr3_+_184529948 | 0.36 |
ENST00000436792.2 ENST00000446204.2 ENST00000422105.1 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr7_+_99699179 | 0.36 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr2_-_105946491 | 0.35 |
ENST00000393359.2 |
TGFBRAP1 |
transforming growth factor, beta receptor associated protein 1 |
| chr8_+_142138711 | 0.35 |
ENST00000518347.1 ENST00000262585.2 ENST00000424248.1 ENST00000519811.1 ENST00000520986.1 ENST00000523058.1 |
DENND3 |
DENN/MADD domain containing 3 |
| chr9_-_74980113 | 0.35 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr17_+_17876127 | 0.34 |
ENST00000582416.1 ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48 |
leucine rich repeat containing 48 |
| chr17_+_46048376 | 0.34 |
ENST00000338399.4 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
| chr1_-_32169920 | 0.34 |
ENST00000373672.3 ENST00000373668.3 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr22_-_20850128 | 0.34 |
ENST00000328879.4 |
KLHL22 |
kelch-like family member 22 |
| chr2_-_242254595 | 0.34 |
ENST00000441124.1 ENST00000391976.2 |
HDLBP |
high density lipoprotein binding protein |
| chr15_-_43785303 | 0.33 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr1_+_160313062 | 0.33 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr1_-_78149041 | 0.32 |
ENST00000414381.1 ENST00000370798.1 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
| chrX_-_102941596 | 0.32 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr16_-_67514982 | 0.32 |
ENST00000565835.1 ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr11_-_64646086 | 0.32 |
ENST00000320631.3 |
EHD1 |
EH-domain containing 1 |
| chr5_-_94890648 | 0.31 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr12_+_54718904 | 0.30 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chr17_-_19771242 | 0.29 |
ENST00000361658.2 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr7_-_6523755 | 0.29 |
ENST00000436575.1 ENST00000258739.4 |
DAGLB KDELR2 |
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr14_-_106092403 | 0.29 |
ENST00000390543.2 |
IGHG4 |
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr16_-_70719925 | 0.29 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr20_+_34359905 | 0.29 |
ENST00000374012.3 ENST00000439301.1 ENST00000339089.6 ENST00000374000.4 |
PHF20 |
PHD finger protein 20 |
| chr3_+_184529929 | 0.28 |
ENST00000287546.4 ENST00000437079.3 |
VPS8 |
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chrX_-_153059958 | 0.28 |
ENST00000370092.3 ENST00000217901.5 |
IDH3G |
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr21_-_46237959 | 0.27 |
ENST00000397898.3 ENST00000411651.2 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr14_-_81687575 | 0.27 |
ENST00000434192.2 |
GTF2A1 |
general transcription factor IIA, 1, 19/37kDa |
| chr12_+_110562135 | 0.27 |
ENST00000361948.4 ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81 |
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr16_+_19729586 | 0.26 |
ENST00000564186.1 ENST00000541926.1 ENST00000433597.2 |
IQCK |
IQ motif containing K |
| chr16_-_30366672 | 0.26 |
ENST00000305596.3 |
CD2BP2 |
CD2 (cytoplasmic tail) binding protein 2 |
| chr12_-_49412588 | 0.26 |
ENST00000547082.1 ENST00000395170.3 |
PRKAG1 |
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr17_+_59529743 | 0.25 |
ENST00000589003.1 ENST00000393853.4 |
TBX4 |
T-box 4 |
| chrX_+_100663243 | 0.25 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr11_-_14521379 | 0.25 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr1_-_78148324 | 0.25 |
ENST00000370801.3 ENST00000433749.1 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
| chr11_+_58910295 | 0.24 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr16_-_2827128 | 0.24 |
ENST00000494946.2 ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2 |
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr1_+_40505891 | 0.23 |
ENST00000372797.3 ENST00000372802.1 ENST00000449311.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr12_-_49412541 | 0.23 |
ENST00000547306.1 ENST00000548857.1 ENST00000551696.1 ENST00000316299.5 |
PRKAG1 |
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr4_-_54930790 | 0.23 |
ENST00000263921.3 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
| chr6_+_127588020 | 0.23 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr15_-_72668185 | 0.23 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr1_+_32645645 | 0.22 |
ENST00000373609.1 |
TXLNA |
taxilin alpha |
| chr9_+_99212403 | 0.22 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr1_+_32645269 | 0.22 |
ENST00000373610.3 |
TXLNA |
taxilin alpha |
| chr9_-_123476612 | 0.22 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr17_-_15466742 | 0.22 |
ENST00000584811.1 ENST00000419890.2 |
TVP23C |
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr9_-_123476719 | 0.22 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr14_+_77787227 | 0.22 |
ENST00000216465.5 ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr17_+_57642886 | 0.22 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr8_-_145016692 | 0.22 |
ENST00000357649.2 |
PLEC |
plectin |
| chr19_-_52227221 | 0.22 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr1_-_85040090 | 0.22 |
ENST00000370630.5 |
CTBS |
chitobiase, di-N-acetyl- |
| chr6_-_25042231 | 0.21 |
ENST00000510784.2 |
FAM65B |
family with sequence similarity 65, member B |
| chr17_+_7211280 | 0.21 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr7_-_27169801 | 0.21 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr8_+_109455845 | 0.21 |
ENST00000220853.3 |
EMC2 |
ER membrane protein complex subunit 2 |
| chr20_+_34043085 | 0.21 |
ENST00000397527.1 ENST00000342580.4 |
CEP250 |
centrosomal protein 250kDa |
| chr8_+_61429416 | 0.21 |
ENST00000262646.7 ENST00000531289.1 |
RAB2A |
RAB2A, member RAS oncogene family |
| chr11_+_118230287 | 0.20 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chrX_-_153059811 | 0.20 |
ENST00000427365.2 ENST00000444450.1 ENST00000370093.1 |
IDH3G |
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr12_-_125398850 | 0.20 |
ENST00000535859.1 ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC |
ubiquitin C |
| chr19_+_2269485 | 0.20 |
ENST00000582888.4 ENST00000602676.2 ENST00000322297.4 ENST00000583542.4 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
| chr8_+_22422749 | 0.20 |
ENST00000523900.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr20_+_2821340 | 0.20 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr3_-_139108475 | 0.20 |
ENST00000515006.1 ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2 |
coatomer protein complex, subunit beta 2 (beta prime) |
| chr5_+_151151504 | 0.19 |
ENST00000356245.3 ENST00000507878.2 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr8_+_97657449 | 0.19 |
ENST00000220763.5 |
CPQ |
carboxypeptidase Q |
| chr17_-_7197881 | 0.19 |
ENST00000007699.5 |
YBX2 |
Y box binding protein 2 |
| chrX_-_102319092 | 0.19 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr12_+_54447637 | 0.19 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr3_+_51428704 | 0.18 |
ENST00000323686.4 |
RBM15B |
RNA binding motif protein 15B |
| chr21_-_38445297 | 0.18 |
ENST00000430792.1 ENST00000399103.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_133787586 | 0.18 |
ENST00000395379.1 ENST00000395386.2 ENST00000337920.4 |
PHF20L1 |
PHD finger protein 20-like 1 |
| chr10_+_103825080 | 0.18 |
ENST00000299238.5 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
| chrX_-_139587225 | 0.17 |
ENST00000370536.2 |
SOX3 |
SRY (sex determining region Y)-box 3 |
| chr16_-_86588627 | 0.17 |
ENST00000565482.1 ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD |
methenyltetrahydrofolate synthetase domain containing |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.8 | 4.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 2.0 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 1.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 1.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 1.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 1.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.6 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.7 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.0 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 4.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 1.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.9 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 8.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 1.3 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.5 | 1.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.5 | 2.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.5 | 1.4 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.3 | 1.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.0 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 5.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 1.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.3 | 1.2 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 1.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.2 | 0.6 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.2 | 2.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 0.8 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 1.8 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 0.8 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.2 | 0.9 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 0.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 0.5 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.2 | 0.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 | 1.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 3.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 2.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.8 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 1.0 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.3 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.1 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.6 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 3.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) specification of axis polarity(GO:0065001) |
| 0.0 | 0.8 | GO:0034329 | cell junction assembly(GO:0034329) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 1.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.3 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0045007 | DNA dealkylation involved in DNA repair(GO:0006307) depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0035655 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0060849 | radial pattern formation(GO:0009956) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 1.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 1.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.4 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.7 | GO:0030818 | negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.0 | 0.1 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 4.2 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.1 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 3.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.8 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.2 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.6 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.7 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.3 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.0 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 4.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |