Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
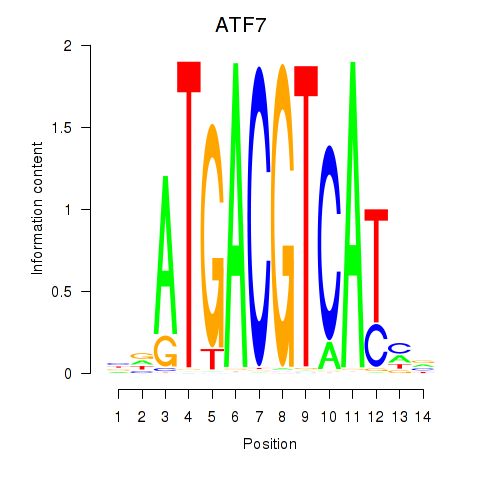
Results for ATF7
Z-value: 0.85
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | ATF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54020107_54020199, hg19_v2_chr12_-_53994805_53994817 | -0.06 | 8.2e-01 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_75311019 | 2.33 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr4_+_75310851 | 2.17 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr17_-_39780819 | 1.60 |
ENST00000311208.8 |
KRT17 |
keratin 17 |
| chr4_+_75480629 | 1.54 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr14_+_68086515 | 1.26 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr7_-_140624499 | 1.21 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr1_+_156030937 | 1.17 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr1_+_26496362 | 1.06 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr6_-_26285737 | 1.02 |
ENST00000377727.1 ENST00000289352.1 |
HIST1H4H |
histone cluster 1, H4h |
| chr16_+_2039946 | 0.97 |
ENST00000248121.2 ENST00000568896.1 |
SYNGR3 |
synaptogyrin 3 |
| chr4_+_85504075 | 0.95 |
ENST00000295887.5 |
CDS1 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr7_-_124569991 | 0.92 |
ENST00000446993.1 ENST00000357628.3 ENST00000393329.1 |
POT1 |
protection of telomeres 1 |
| chr3_+_158288999 | 0.84 |
ENST00000482628.1 ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr7_+_5632436 | 0.84 |
ENST00000340250.6 ENST00000382361.3 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr6_-_31774714 | 0.79 |
ENST00000375661.5 |
LSM2 |
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_+_7155556 | 0.77 |
ENST00000570500.1 ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr16_-_3767506 | 0.74 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr3_+_158288942 | 0.74 |
ENST00000491767.1 ENST00000355893.5 |
MLF1 |
myeloid leukemia factor 1 |
| chr3_+_158288960 | 0.72 |
ENST00000484955.1 ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr10_+_22634384 | 0.72 |
ENST00000376624.3 ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6 |
sperm associated antigen 6 |
| chr4_-_122744998 | 0.72 |
ENST00000274026.5 |
CCNA2 |
cyclin A2 |
| chr11_-_28129656 | 0.68 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr5_-_16509101 | 0.67 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr16_-_3767551 | 0.66 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr11_+_3819049 | 0.61 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chrX_+_150148976 | 0.59 |
ENST00000419110.1 |
HMGB3 |
high mobility group box 3 |
| chr10_+_35484793 | 0.59 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr11_-_77185094 | 0.58 |
ENST00000278568.4 ENST00000356341.3 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr17_-_8151353 | 0.58 |
ENST00000315684.8 |
CTC1 |
CTS telomere maintenance complex component 1 |
| chr11_-_3818688 | 0.57 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr7_+_23221613 | 0.56 |
ENST00000410002.3 ENST00000413919.1 |
NUPL2 |
nucleoporin like 2 |
| chr11_+_17281900 | 0.56 |
ENST00000530527.1 |
NUCB2 |
nucleobindin 2 |
| chrX_+_110339439 | 0.55 |
ENST00000372010.1 ENST00000519681.1 ENST00000372007.5 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr17_-_49124230 | 0.55 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr19_+_36486078 | 0.55 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr1_+_2004901 | 0.55 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr6_+_28048753 | 0.55 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr5_-_175964366 | 0.55 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr4_+_128802016 | 0.54 |
ENST00000270861.5 ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4 |
polo-like kinase 4 |
| chr19_+_10362577 | 0.54 |
ENST00000592514.1 ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr9_-_33001520 | 0.53 |
ENST00000463596.1 ENST00000379819.1 ENST00000397172.3 ENST00000379812.5 ENST00000477119.1 ENST00000379813.3 ENST00000379825.2 ENST00000309615.3 ENST00000476858.1 ENST00000473221.1 |
APTX |
aprataxin |
| chr11_+_18343800 | 0.53 |
ENST00000453096.2 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr19_+_10362882 | 0.52 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr1_+_39456895 | 0.51 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr16_+_81040794 | 0.51 |
ENST00000439957.3 ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN |
centromere protein N |
| chr4_+_170581213 | 0.49 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr13_-_31736478 | 0.48 |
ENST00000445273.2 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr15_-_91537723 | 0.48 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr16_+_68056844 | 0.48 |
ENST00000565263.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr16_+_68057153 | 0.47 |
ENST00000358896.6 ENST00000568099.2 |
DUS2 |
dihydrouridine synthase 2 |
| chr11_-_123525289 | 0.47 |
ENST00000392770.2 ENST00000299333.3 ENST00000530277.1 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
| chr7_+_23221438 | 0.47 |
ENST00000258742.5 |
NUPL2 |
nucleoporin like 2 |
| chr16_+_68057179 | 0.46 |
ENST00000567100.1 ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2 |
dihydrouridine synthase 2 |
| chr17_+_7155343 | 0.46 |
ENST00000573513.1 ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr9_-_131644306 | 0.46 |
ENST00000302586.3 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr7_-_73153178 | 0.45 |
ENST00000437775.2 ENST00000222800.3 |
ABHD11 |
abhydrolase domain containing 11 |
| chr9_-_131644202 | 0.44 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr11_-_62457371 | 0.43 |
ENST00000317449.4 |
LRRN4CL |
LRRN4 C-terminal like |
| chr22_+_32340447 | 0.43 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr19_-_8008533 | 0.43 |
ENST00000597926.1 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr19_+_44100632 | 0.42 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
| chr7_+_56019486 | 0.42 |
ENST00000446692.1 ENST00000285298.4 ENST00000443449.1 |
GBAS MRPS17 |
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr21_+_45209394 | 0.40 |
ENST00000497547.1 |
RRP1 |
ribosomal RNA processing 1 |
| chr1_+_2005425 | 0.40 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr16_-_81040719 | 0.40 |
ENST00000219400.3 |
CMC2 |
C-x(9)-C motif containing 2 |
| chr1_-_47184745 | 0.39 |
ENST00000544071.1 |
EFCAB14 |
EF-hand calcium binding domain 14 |
| chr12_+_132628963 | 0.39 |
ENST00000330579.1 |
NOC4L |
nucleolar complex associated 4 homolog (S. cerevisiae) |
| chr8_+_98788003 | 0.39 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr7_+_100464760 | 0.38 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr3_-_33759699 | 0.38 |
ENST00000399362.4 ENST00000359576.5 ENST00000307312.7 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr17_+_34431212 | 0.37 |
ENST00000394495.1 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr22_+_32340481 | 0.37 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr6_-_32557610 | 0.37 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr20_+_42295745 | 0.37 |
ENST00000396863.4 ENST00000217026.4 |
MYBL2 |
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr13_+_111767650 | 0.37 |
ENST00000449979.1 ENST00000370623.3 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_-_146019693 | 0.37 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr15_-_42840961 | 0.37 |
ENST00000563454.1 ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57 |
leucine rich repeat containing 57 |
| chr6_-_27440460 | 0.36 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chr11_-_18343669 | 0.36 |
ENST00000396253.3 ENST00000349215.3 ENST00000438420.2 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
| chr19_+_44100727 | 0.35 |
ENST00000528387.1 ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576 SRRM5 |
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr14_-_101295407 | 0.35 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr14_+_60716159 | 0.35 |
ENST00000325658.3 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr12_-_51566562 | 0.35 |
ENST00000548108.1 |
TFCP2 |
transcription factor CP2 |
| chr10_+_119000604 | 0.35 |
ENST00000298472.5 |
SLC18A2 |
solute carrier family 18 (vesicular monoamine transporter), member 2 |
| chr14_-_102552659 | 0.34 |
ENST00000441629.2 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chrX_-_7895755 | 0.34 |
ENST00000444736.1 ENST00000537427.1 ENST00000442940.1 |
PNPLA4 |
patatin-like phospholipase domain containing 4 |
| chr4_+_108910870 | 0.34 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr10_+_35484053 | 0.33 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr14_+_60715928 | 0.33 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr11_+_108535752 | 0.33 |
ENST00000322536.3 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr4_+_108911036 | 0.33 |
ENST00000505878.1 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr6_-_27440837 | 0.32 |
ENST00000211936.6 |
ZNF184 |
zinc finger protein 184 |
| chr3_-_33759541 | 0.32 |
ENST00000468888.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr19_+_36036583 | 0.32 |
ENST00000392205.1 |
TMEM147 |
transmembrane protein 147 |
| chr4_-_668108 | 0.32 |
ENST00000304312.4 |
ATP5I |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr19_-_47616992 | 0.32 |
ENST00000253048.5 |
ZC3H4 |
zinc finger CCCH-type containing 4 |
| chr7_+_152456829 | 0.32 |
ENST00000377776.3 ENST00000256001.8 ENST00000397282.2 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
| chr6_+_37400974 | 0.31 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr11_+_4116005 | 0.30 |
ENST00000300738.5 |
RRM1 |
ribonucleotide reductase M1 |
| chr12_-_93835665 | 0.30 |
ENST00000552442.1 ENST00000550657.1 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
| chr12_-_51566849 | 0.30 |
ENST00000549867.1 ENST00000307660.4 |
TFCP2 |
transcription factor CP2 |
| chr6_-_34855773 | 0.30 |
ENST00000420584.2 ENST00000361288.4 |
TAF11 |
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr6_-_26032288 | 0.29 |
ENST00000244661.2 |
HIST1H3B |
histone cluster 1, H3b |
| chr4_-_2010562 | 0.29 |
ENST00000411649.1 ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA |
negative elongation factor complex member A |
| chr19_+_59055814 | 0.28 |
ENST00000594806.1 ENST00000253024.5 ENST00000341753.6 |
TRIM28 |
tripartite motif containing 28 |
| chr3_+_170075436 | 0.28 |
ENST00000476188.1 ENST00000259119.4 ENST00000426052.2 |
SKIL |
SKI-like oncogene |
| chr4_-_146019287 | 0.28 |
ENST00000502847.1 ENST00000513054.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr9_-_111696224 | 0.28 |
ENST00000537196.1 |
IKBKAP |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr11_+_4116054 | 0.28 |
ENST00000423050.2 |
RRM1 |
ribonucleotide reductase M1 |
| chr4_+_1873100 | 0.27 |
ENST00000508803.1 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
| chr1_-_1284730 | 0.27 |
ENST00000378888.5 |
DVL1 |
dishevelled segment polarity protein 1 |
| chr11_-_3818932 | 0.27 |
ENST00000324932.7 ENST00000359171.4 |
NUP98 |
nucleoporin 98kDa |
| chr22_-_44258360 | 0.27 |
ENST00000330884.4 ENST00000249130.5 |
SULT4A1 |
sulfotransferase family 4A, member 1 |
| chr4_-_104119528 | 0.27 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chr9_+_70856899 | 0.27 |
ENST00000377342.5 ENST00000478048.1 |
CBWD3 |
COBW domain containing 3 |
| chrX_+_77359671 | 0.27 |
ENST00000373316.4 |
PGK1 |
phosphoglycerate kinase 1 |
| chr16_+_56225248 | 0.26 |
ENST00000262493.6 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr2_+_74781828 | 0.26 |
ENST00000340004.6 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr18_+_72163443 | 0.26 |
ENST00000324262.4 ENST00000580672.1 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr12_-_51566592 | 0.26 |
ENST00000257915.5 ENST00000548115.1 |
TFCP2 |
transcription factor CP2 |
| chr22_+_31518938 | 0.26 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr17_+_34430980 | 0.26 |
ENST00000250151.4 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr12_+_112856690 | 0.26 |
ENST00000392597.1 ENST00000351677.2 |
PTPN11 |
protein tyrosine phosphatase, non-receptor type 11 |
| chr9_-_69262509 | 0.25 |
ENST00000377449.1 ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6 |
COBW domain containing 6 |
| chr2_-_43823119 | 0.24 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr19_+_35861831 | 0.24 |
ENST00000454971.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr9_-_179018 | 0.24 |
ENST00000431099.2 ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1 |
COBW domain containing 1 |
| chr6_-_106773610 | 0.24 |
ENST00000369076.3 ENST00000369070.1 |
ATG5 |
autophagy related 5 |
| chr18_+_72163536 | 0.23 |
ENST00000579847.1 ENST00000583203.1 ENST00000581513.1 ENST00000577600.1 ENST00000579583.1 ENST00000584613.1 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_-_146019335 | 0.23 |
ENST00000451299.2 ENST00000507656.1 ENST00000309439.5 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr2_-_43823093 | 0.23 |
ENST00000405006.4 |
THADA |
thyroid adenoma associated |
| chrX_+_77359726 | 0.23 |
ENST00000442431.1 |
PGK1 |
phosphoglycerate kinase 1 |
| chr11_+_18344106 | 0.23 |
ENST00000534641.1 ENST00000525831.1 ENST00000265963.4 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr19_-_48547294 | 0.23 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chr3_+_101292939 | 0.23 |
ENST00000265260.3 ENST00000469941.1 ENST00000296024.5 |
PCNP |
PEST proteolytic signal containing nuclear protein |
| chr9_-_70490107 | 0.22 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr5_+_131892603 | 0.22 |
ENST00000378823.3 ENST00000265335.6 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
| chr12_+_57998400 | 0.22 |
ENST00000548804.1 ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr18_+_2571510 | 0.22 |
ENST00000261597.4 ENST00000575515.1 |
NDC80 |
NDC80 kinetochore complex component |
| chr19_+_36103631 | 0.22 |
ENST00000203166.5 ENST00000379045.2 |
HAUS5 |
HAUS augmin-like complex, subunit 5 |
| chrX_+_155110956 | 0.21 |
ENST00000286448.6 ENST00000262640.6 ENST00000460621.1 |
VAMP7 |
vesicle-associated membrane protein 7 |
| chr16_-_3068171 | 0.21 |
ENST00000572154.1 ENST00000328796.4 |
CLDN6 |
claudin 6 |
| chr2_+_114195268 | 0.21 |
ENST00000259199.4 ENST00000416503.2 ENST00000433343.2 |
CBWD2 |
COBW domain containing 2 |
| chr5_+_112849373 | 0.21 |
ENST00000161863.4 ENST00000515883.1 |
YTHDC2 |
YTH domain containing 2 |
| chr7_-_44924939 | 0.21 |
ENST00000395699.2 |
PURB |
purine-rich element binding protein B |
| chr12_-_93836028 | 0.21 |
ENST00000318066.2 |
UBE2N |
ubiquitin-conjugating enzyme E2N |
| chr17_+_43209967 | 0.21 |
ENST00000431281.1 ENST00000591859.1 |
ACBD4 |
acyl-CoA binding domain containing 4 |
| chr17_+_29421900 | 0.21 |
ENST00000358273.4 ENST00000356175.3 |
NF1 |
neurofibromin 1 |
| chr17_-_685559 | 0.20 |
ENST00000301329.6 |
GLOD4 |
glyoxalase domain containing 4 |
| chr5_-_138210977 | 0.20 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr12_+_56521840 | 0.20 |
ENST00000394048.5 |
ESYT1 |
extended synaptotagmin-like protein 1 |
| chr19_+_45504688 | 0.20 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr1_-_22109682 | 0.20 |
ENST00000400301.1 ENST00000532737.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr18_+_76829441 | 0.20 |
ENST00000458297.2 |
ATP9B |
ATPase, class II, type 9B |
| chr17_+_41177220 | 0.20 |
ENST00000587250.2 ENST00000544533.1 |
RND2 |
Rho family GTPase 2 |
| chr3_+_38206975 | 0.20 |
ENST00000446845.1 ENST00000311806.3 |
OXSR1 |
oxidative stress responsive 1 |
| chr8_+_26149007 | 0.20 |
ENST00000380737.3 ENST00000524169.1 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr19_+_1941117 | 0.20 |
ENST00000255641.8 |
CSNK1G2 |
casein kinase 1, gamma 2 |
| chr15_+_42841008 | 0.19 |
ENST00000260372.3 ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2 |
HAUS augmin-like complex, subunit 2 |
| chr1_-_202896310 | 0.19 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr22_-_19132154 | 0.19 |
ENST00000252137.6 |
DGCR14 |
DiGeorge syndrome critical region gene 14 |
| chr19_-_460996 | 0.19 |
ENST00000264554.6 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr20_+_5892037 | 0.19 |
ENST00000378961.4 |
CHGB |
chromogranin B (secretogranin 1) |
| chr19_+_35862192 | 0.19 |
ENST00000597214.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr12_+_57998595 | 0.19 |
ENST00000337737.3 ENST00000548198.1 ENST00000551632.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr2_-_220174166 | 0.18 |
ENST00000409251.3 ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr9_+_70856397 | 0.18 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr11_-_62389621 | 0.18 |
ENST00000531383.1 ENST00000265471.5 |
B3GAT3 |
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr6_-_144416737 | 0.18 |
ENST00000367569.2 |
SF3B5 |
splicing factor 3b, subunit 5, 10kDa |
| chr19_+_13056663 | 0.18 |
ENST00000541222.1 ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
| chr22_+_25003626 | 0.18 |
ENST00000451366.1 ENST00000406383.2 ENST00000428855.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr12_+_100660909 | 0.18 |
ENST00000549687.1 |
SCYL2 |
SCY1-like 2 (S. cerevisiae) |
| chr17_-_685493 | 0.17 |
ENST00000536578.1 ENST00000301328.5 ENST00000576419.1 |
GLOD4 |
glyoxalase domain containing 4 |
| chr6_-_42713792 | 0.17 |
ENST00000372876.1 |
TBCC |
tubulin folding cofactor C |
| chr14_+_105219437 | 0.17 |
ENST00000329967.6 ENST00000347067.5 ENST00000553810.1 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
| chr2_-_220264703 | 0.17 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr1_+_70820451 | 0.17 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr17_+_53342311 | 0.16 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chrX_+_152224766 | 0.16 |
ENST00000370265.4 ENST00000447306.1 |
PNMA3 |
paraneoplastic Ma antigen 3 |
| chr18_+_23806437 | 0.16 |
ENST00000578121.1 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr6_+_32939964 | 0.16 |
ENST00000607833.1 |
BRD2 |
bromodomain containing 2 |
| chr19_-_56826157 | 0.16 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr19_+_18723660 | 0.16 |
ENST00000262817.3 |
TMEM59L |
transmembrane protein 59-like |
| chr14_+_100705322 | 0.16 |
ENST00000262238.4 |
YY1 |
YY1 transcription factor |
| chr3_+_140660743 | 0.16 |
ENST00000453248.2 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr14_+_93651296 | 0.16 |
ENST00000283534.4 ENST00000557574.1 |
TMEM251 RP11-371E8.4 |
transmembrane protein 251 Uncharacterized protein |
| chr14_+_93651358 | 0.15 |
ENST00000415050.2 |
TMEM251 |
transmembrane protein 251 |
| chr11_+_82783097 | 0.15 |
ENST00000501011.2 ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1 |
RAB30 antisense RNA 1 (head to head) |
| chr11_-_8680383 | 0.15 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr18_-_2571437 | 0.15 |
ENST00000574676.1 ENST00000574538.1 ENST00000319888.6 |
METTL4 |
methyltransferase like 4 |
| chr19_+_35849723 | 0.14 |
ENST00000594310.1 |
FFAR3 |
free fatty acid receptor 3 |
| chr2_-_97760576 | 0.14 |
ENST00000414820.1 ENST00000272610.3 |
FAHD2B |
fumarylacetoacetate hydrolase domain containing 2B |
| chr3_-_21792838 | 0.14 |
ENST00000281523.2 |
ZNF385D |
zinc finger protein 385D |
| chr12_+_100661156 | 0.14 |
ENST00000360820.2 |
SCYL2 |
SCY1-like 2 (S. cerevisiae) |
| chr19_-_40919271 | 0.14 |
ENST00000291825.7 ENST00000324001.7 |
PRX |
periaxin |
| chr19_-_10341948 | 0.14 |
ENST00000590320.1 ENST00000592342.1 ENST00000588952.1 |
S1PR2 DNMT1 |
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr1_+_168148169 | 0.14 |
ENST00000367833.2 |
TIPRL |
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr6_-_16761678 | 0.14 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr6_-_106773491 | 0.13 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr10_-_135122346 | 0.13 |
ENST00000417178.2 |
TUBGCP2 |
tubulin, gamma complex associated protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.4 | 3.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 1.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.2 | 0.7 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.2 | 1.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.2 | 0.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 1.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.9 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 1.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.3 | GO:0043400 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.1 | 0.5 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.3 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 1.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 1.6 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.3 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.5 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:1901535 | convergent extension involved in axis elongation(GO:0060028) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.5 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.5 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.9 | GO:0051096 | telomere assembly(GO:0032202) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.6 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.6 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 | 0.5 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.4 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.1 | 1.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.2 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.2 | GO:0031860 | regulation of mitotic recombination(GO:0000019) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.9 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.1 | GO:0090309 | maintenance of DNA methylation(GO:0010216) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.4 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 1.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.7 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.0 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.3 | 1.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 0.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 0.5 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 1.7 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 4.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 1.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.5 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 1.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 5.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |