Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
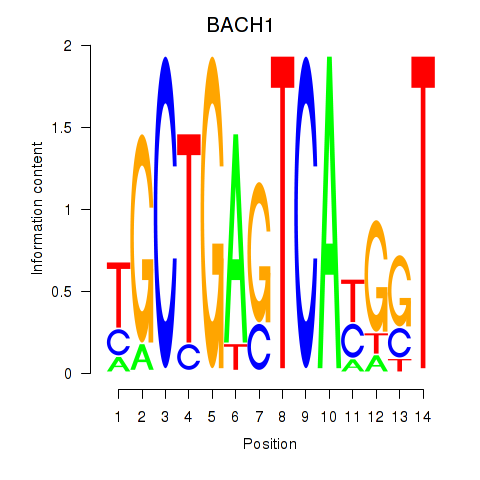
Results for BACH1_NFE2_NFE2L2
Z-value: 1.91
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BACH1 |
|
NFE2
|
ENSG00000123405.9 | NFE2 |
|
NFE2L2
|
ENSG00000116044.11 | NFE2L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2 | hg19_v2_chr12_-_54689532_54689551, hg19_v2_chr12_-_54694807_54694905, hg19_v2_chr12_-_54694758_54694805 | 0.81 | 1.6e-04 | Click! |
| NFE2L2 | hg19_v2_chr2_-_178128250_178128300, hg19_v2_chr2_-_178257401_178257432, hg19_v2_chr2_-_178128528_178128574, hg19_v2_chr2_-_178129853_178129860 | -0.59 | 1.5e-02 | Click! |
| BACH1 | hg19_v2_chr21_+_30671189_30671207, hg19_v2_chr21_+_30671690_30671762 | 0.39 | 1.3e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_154563889 | 8.17 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr8_-_30585217 | 5.26 |
ENST00000520888.1 ENST00000414019.1 |
GSR |
glutathione reductase |
| chr8_-_30585294 | 5.13 |
ENST00000546342.1 ENST00000541648.1 ENST00000537535.1 |
GSR |
glutathione reductase |
| chr8_-_30585439 | 5.11 |
ENST00000221130.5 |
GSR |
glutathione reductase |
| chr2_-_220083076 | 4.17 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr1_-_158656488 | 3.76 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr22_+_19466980 | 3.71 |
ENST00000407835.1 ENST00000438587.1 |
CDC45 |
cell division cycle 45 |
| chr7_+_112063192 | 3.62 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr17_+_30771279 | 3.44 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr1_-_225616515 | 3.10 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr6_-_35888905 | 2.71 |
ENST00000510290.1 ENST00000423325.2 ENST00000373822.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr6_+_32121218 | 2.63 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_+_44214824 | 2.48 |
ENST00000371646.5 ENST00000353801.3 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr5_-_137911049 | 2.46 |
ENST00000297185.3 |
HSPA9 |
heat shock 70kDa protein 9 (mortalin) |
| chr10_-_6019984 | 2.25 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr14_+_58711539 | 2.02 |
ENST00000216455.4 ENST00000412908.2 ENST00000557508.1 |
PSMA3 |
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr16_-_1429627 | 1.96 |
ENST00000248104.7 |
UNKL |
unkempt family zinc finger-like |
| chr10_-_76868931 | 1.90 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr17_-_79881408 | 1.84 |
ENST00000392366.3 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr6_+_44215603 | 1.80 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr10_-_6019552 | 1.80 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr17_-_28618948 | 1.78 |
ENST00000261714.6 |
BLMH |
bleomycin hydrolase |
| chr18_-_55253989 | 1.77 |
ENST00000262093.5 |
FECH |
ferrochelatase |
| chr6_-_35888824 | 1.61 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr16_-_28222797 | 1.60 |
ENST00000569951.1 ENST00000565698.1 |
XPO6 |
exportin 6 |
| chr21_+_33031935 | 1.55 |
ENST00000270142.6 ENST00000389995.4 |
SOD1 |
superoxide dismutase 1, soluble |
| chr11_+_46402297 | 1.53 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr2_-_220173685 | 1.51 |
ENST00000423636.2 ENST00000442029.1 ENST00000412847.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr17_-_28618867 | 1.51 |
ENST00000394819.3 ENST00000577623.1 |
BLMH |
bleomycin hydrolase |
| chr11_+_118958689 | 1.48 |
ENST00000535253.1 ENST00000392841.1 |
HMBS |
hydroxymethylbilane synthase |
| chr19_-_59023348 | 1.40 |
ENST00000601355.1 ENST00000263093.2 |
SLC27A5 |
solute carrier family 27 (fatty acid transporter), member 5 |
| chr17_-_73150629 | 1.36 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
| chr17_+_76311791 | 1.33 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr14_+_103801140 | 1.30 |
ENST00000561325.1 ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chrX_+_153770421 | 1.29 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_-_200992827 | 1.28 |
ENST00000332129.2 ENST00000422435.2 |
KIF21B |
kinesin family member 21B |
| chr12_-_71182695 | 1.26 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr16_-_1429674 | 1.22 |
ENST00000403703.1 ENST00000397464.1 ENST00000402641.2 |
UNKL |
unkempt family zinc finger-like |
| chr16_+_30077055 | 1.22 |
ENST00000564595.2 ENST00000569798.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr3_-_48057890 | 1.13 |
ENST00000434267.1 |
MAP4 |
microtubule-associated protein 4 |
| chr2_-_11606275 | 1.12 |
ENST00000381525.3 ENST00000362009.4 |
E2F6 |
E2F transcription factor 6 |
| chr1_-_150208291 | 1.11 |
ENST00000533654.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_25756638 | 1.09 |
ENST00000349320.3 |
RHCE |
Rh blood group, CcEe antigens |
| chr10_+_45495898 | 1.05 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr8_-_38386175 | 1.02 |
ENST00000437935.2 ENST00000358138.1 |
C8orf86 |
chromosome 8 open reading frame 86 |
| chr7_+_22766766 | 1.01 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr15_-_83240507 | 1.00 |
ENST00000564522.1 ENST00000398592.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr3_-_98241358 | 1.00 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr15_-_83240553 | 0.98 |
ENST00000423133.2 ENST00000398591.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_150208320 | 0.97 |
ENST00000534220.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_62474803 | 0.96 |
ENST00000533982.1 ENST00000360796.5 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_+_184037466 | 0.96 |
ENST00000441154.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_150208363 | 0.94 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_+_184038073 | 0.93 |
ENST00000428387.1 ENST00000434061.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_+_36410762 | 0.93 |
ENST00000483557.1 ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20 |
potassium channel tetramerization domain containing 20 |
| chrX_-_15511438 | 0.92 |
ENST00000380420.5 |
PIR |
pirin (iron-binding nuclear protein) |
| chr7_+_130794846 | 0.92 |
ENST00000421797.2 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
| chr19_-_10613421 | 0.91 |
ENST00000393623.2 |
KEAP1 |
kelch-like ECH-associated protein 1 |
| chr3_+_183903811 | 0.91 |
ENST00000429586.2 ENST00000292808.5 |
ABCF3 |
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr19_-_46285646 | 0.88 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr12_+_64845660 | 0.88 |
ENST00000331710.5 |
TBK1 |
TANK-binding kinase 1 |
| chr18_-_55253871 | 0.87 |
ENST00000382873.3 |
FECH |
ferrochelatase |
| chr22_-_19466683 | 0.87 |
ENST00000399523.1 ENST00000421968.2 ENST00000447868.1 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr1_-_109968973 | 0.84 |
ENST00000271308.4 ENST00000538610.1 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr11_-_9025541 | 0.83 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr17_-_42276574 | 0.83 |
ENST00000589805.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr1_-_150208412 | 0.78 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_14541872 | 0.77 |
ENST00000419365.2 ENST00000530457.1 ENST00000532256.1 ENST00000533068.1 |
PSMA1 |
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr1_-_150208498 | 0.73 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr10_+_35484793 | 0.73 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr22_-_19466732 | 0.73 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr16_-_23521710 | 0.72 |
ENST00000562117.1 ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr4_+_170581213 | 0.72 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr1_+_165796753 | 0.71 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr16_+_12059091 | 0.71 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr8_-_59572093 | 0.71 |
ENST00000427130.2 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr22_+_18893736 | 0.71 |
ENST00000331444.6 |
DGCR6 |
DiGeorge syndrome critical region gene 6 |
| chr13_-_67802549 | 0.71 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr17_+_55162453 | 0.70 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr17_-_61905005 | 0.70 |
ENST00000584574.1 ENST00000585145.1 ENST00000427159.2 |
FTSJ3 |
FtsJ homolog 3 (E. coli) |
| chr10_+_35484053 | 0.70 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr10_+_85899196 | 0.69 |
ENST00000372134.3 |
GHITM |
growth hormone inducible transmembrane protein |
| chr7_+_100026406 | 0.68 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chrX_-_16730688 | 0.68 |
ENST00000359276.4 |
CTPS2 |
CTP synthase 2 |
| chr7_+_101460882 | 0.68 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr14_+_103589789 | 0.68 |
ENST00000558056.1 ENST00000560869.1 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
| chr16_+_30211181 | 0.67 |
ENST00000395138.2 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr3_+_69915385 | 0.67 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr16_+_22308717 | 0.66 |
ENST00000299853.5 ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr22_-_32058166 | 0.65 |
ENST00000435900.1 ENST00000336566.4 |
PISD |
phosphatidylserine decarboxylase |
| chr6_-_31704282 | 0.64 |
ENST00000375784.3 ENST00000375779.2 |
CLIC1 |
chloride intracellular channel 1 |
| chr1_+_76251879 | 0.64 |
ENST00000535300.1 ENST00000319942.3 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
| chr12_-_15114492 | 0.63 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_12677714 | 0.63 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr6_-_3157760 | 0.63 |
ENST00000333628.3 |
TUBB2A |
tubulin, beta 2A class IIa |
| chrX_-_16730984 | 0.63 |
ENST00000380241.3 |
CTPS2 |
CTP synthase 2 |
| chr17_+_61905058 | 0.62 |
ENST00000375812.4 ENST00000581882.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr16_+_12059050 | 0.62 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_926000 | 0.61 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr9_+_139921916 | 0.61 |
ENST00000314330.2 |
C9orf139 |
chromosome 9 open reading frame 139 |
| chr5_+_118690466 | 0.60 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr2_+_122513109 | 0.58 |
ENST00000389682.3 ENST00000536142.1 |
TSN |
translin |
| chr1_-_39339777 | 0.58 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chrX_-_109683446 | 0.56 |
ENST00000372057.1 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr6_+_84569359 | 0.56 |
ENST00000369681.5 ENST00000369679.4 |
CYB5R4 |
cytochrome b5 reductase 4 |
| chr20_+_60878005 | 0.55 |
ENST00000253003.2 |
ADRM1 |
adhesion regulating molecule 1 |
| chr14_-_95942173 | 0.55 |
ENST00000334258.5 ENST00000557275.1 ENST00000553340.1 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
| chr8_-_59572301 | 0.54 |
ENST00000038176.3 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr4_-_987217 | 0.54 |
ENST00000361661.2 ENST00000398516.2 |
SLC26A1 |
solute carrier family 26 (anion exchanger), member 1 |
| chr11_-_9781068 | 0.53 |
ENST00000500698.1 |
RP11-540A21.2 |
RP11-540A21.2 |
| chr10_-_94333784 | 0.53 |
ENST00000265986.6 |
IDE |
insulin-degrading enzyme |
| chr11_-_2924970 | 0.53 |
ENST00000533594.1 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr8_-_82395461 | 0.53 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr16_+_29819446 | 0.52 |
ENST00000568282.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_-_15180257 | 0.50 |
ENST00000540462.1 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr20_+_34742650 | 0.50 |
ENST00000373945.1 ENST00000338074.2 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr22_+_23165153 | 0.50 |
ENST00000390317.2 |
IGLV2-8 |
immunoglobulin lambda variable 2-8 |
| chr18_+_21693306 | 0.49 |
ENST00000540918.2 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr6_+_33378738 | 0.49 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chrX_+_99899180 | 0.48 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr15_+_78833071 | 0.48 |
ENST00000559365.1 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr17_+_79935464 | 0.48 |
ENST00000581647.1 ENST00000580534.1 ENST00000579684.1 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_+_63448918 | 0.47 |
ENST00000341307.2 ENST00000356000.3 ENST00000542238.1 |
RTN3 |
reticulon 3 |
| chr1_+_17634689 | 0.47 |
ENST00000375453.1 ENST00000375448.4 |
PADI4 |
peptidyl arginine deiminase, type IV |
| chr6_-_39902160 | 0.45 |
ENST00000340692.5 |
MOCS1 |
molybdenum cofactor synthesis 1 |
| chr16_+_30077098 | 0.45 |
ENST00000395240.3 ENST00000566846.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr7_-_129845188 | 0.45 |
ENST00000462753.1 ENST00000471077.1 ENST00000473456.1 ENST00000336804.8 |
TMEM209 |
transmembrane protein 209 |
| chr11_-_47447767 | 0.45 |
ENST00000530651.1 ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chrX_+_22056165 | 0.44 |
ENST00000535894.1 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr9_+_116267536 | 0.44 |
ENST00000374136.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr15_+_78832747 | 0.44 |
ENST00000560217.1 ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4 |
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr11_+_10477733 | 0.44 |
ENST00000528723.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr19_+_18496957 | 0.44 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr12_-_26986076 | 0.44 |
ENST00000381340.3 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr4_-_146019693 | 0.43 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr5_+_179247759 | 0.43 |
ENST00000389805.4 ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1 |
sequestosome 1 |
| chr7_+_102988082 | 0.43 |
ENST00000292644.3 ENST00000544811.1 |
PSMC2 |
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
| chr5_+_179125368 | 0.42 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr13_-_41593425 | 0.42 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr17_+_18759612 | 0.42 |
ENST00000432893.2 ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2 |
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr6_-_28321971 | 0.42 |
ENST00000396838.2 ENST00000426434.1 ENST00000434036.1 ENST00000439628.1 |
ZSCAN31 |
zinc finger and SCAN domain containing 31 |
| chr19_-_6670128 | 0.41 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr2_+_231921574 | 0.41 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr1_+_14075865 | 0.41 |
ENST00000413440.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr7_-_115670804 | 0.40 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr2_+_74781828 | 0.40 |
ENST00000340004.6 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr5_-_175964366 | 0.40 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr4_+_17616253 | 0.39 |
ENST00000237380.7 |
MED28 |
mediator complex subunit 28 |
| chr22_-_31063782 | 0.39 |
ENST00000404885.1 ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18 |
dual specificity phosphatase 18 |
| chr1_+_14075903 | 0.39 |
ENST00000343137.4 ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr11_-_65430251 | 0.39 |
ENST00000534283.1 ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA |
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr1_+_100111580 | 0.39 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chr22_+_22681656 | 0.38 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr20_-_48782639 | 0.38 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr15_+_41851211 | 0.38 |
ENST00000263798.3 |
TYRO3 |
TYRO3 protein tyrosine kinase |
| chr22_+_22730353 | 0.38 |
ENST00000390296.2 |
IGLV5-45 |
immunoglobulin lambda variable 5-45 |
| chr8_-_95220775 | 0.37 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr6_-_31745085 | 0.37 |
ENST00000375686.3 ENST00000447450.1 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr8_-_94753229 | 0.36 |
ENST00000518597.1 ENST00000399300.2 ENST00000517700.1 |
RBM12B |
RNA binding motif protein 12B |
| chr4_-_26492076 | 0.36 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr7_+_150434430 | 0.36 |
ENST00000358647.3 |
GIMAP5 |
GTPase, IMAP family member 5 |
| chr7_-_115670792 | 0.36 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr19_-_49658387 | 0.36 |
ENST00000595625.1 |
HRC |
histidine rich calcium binding protein |
| chr6_-_31745037 | 0.36 |
ENST00000375688.4 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr9_-_116840728 | 0.36 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr17_+_61904766 | 0.35 |
ENST00000581842.1 ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr11_-_47447970 | 0.35 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr2_-_89417335 | 0.35 |
ENST00000490686.1 |
IGKV1-17 |
immunoglobulin kappa variable 1-17 |
| chr3_-_119276575 | 0.35 |
ENST00000383669.3 ENST00000383668.3 |
CD80 |
CD80 molecule |
| chr11_+_125365110 | 0.35 |
ENST00000527818.1 |
AP000708.1 |
AP000708.1 |
| chr12_-_15114603 | 0.35 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_156756667 | 0.35 |
ENST00000526188.1 ENST00000454659.1 |
PRCC |
papillary renal cell carcinoma (translocation-associated) |
| chr4_+_123747834 | 0.34 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr6_-_39902185 | 0.34 |
ENST00000373195.3 ENST00000308559.7 ENST00000373188.2 |
MOCS1 |
molybdenum cofactor synthesis 1 |
| chr18_-_52989217 | 0.34 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr17_-_74733404 | 0.34 |
ENST00000508921.3 ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2 |
serine/arginine-rich splicing factor 2 |
| chr22_+_39353527 | 0.33 |
ENST00000249116.2 |
APOBEC3A |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr6_-_166582107 | 0.33 |
ENST00000296946.2 ENST00000461348.2 ENST00000366871.3 |
T |
T, brachyury homolog (mouse) |
| chr2_-_89385283 | 0.33 |
ENST00000390252.2 |
IGKV3-15 |
immunoglobulin kappa variable 3-15 |
| chr3_-_98241760 | 0.33 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chr10_-_21786179 | 0.32 |
ENST00000377113.5 |
CASC10 |
cancer susceptibility candidate 10 |
| chr12_-_113909877 | 0.32 |
ENST00000261731.3 |
LHX5 |
LIM homeobox 5 |
| chrX_-_119445306 | 0.31 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr4_+_120056939 | 0.31 |
ENST00000307128.5 |
MYOZ2 |
myozenin 2 |
| chr11_-_2906979 | 0.30 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr14_+_103800513 | 0.30 |
ENST00000560338.1 ENST00000560763.1 ENST00000558316.1 ENST00000558265.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chr9_+_141107506 | 0.30 |
ENST00000446912.2 |
FAM157B |
family with sequence similarity 157, member B |
| chr7_-_102985035 | 0.30 |
ENST00000426036.2 ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2 |
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr2_-_169887827 | 0.30 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr12_-_53343602 | 0.30 |
ENST00000546897.1 ENST00000552551.1 |
KRT8 |
keratin 8 |
| chr11_+_67007518 | 0.30 |
ENST00000530342.1 ENST00000308783.5 |
KDM2A |
lysine (K)-specific demethylase 2A |
| chr6_+_87646995 | 0.30 |
ENST00000305344.5 |
HTR1E |
5-hydroxytryptamine (serotonin) receptor 1E, G protein-coupled |
| chr11_-_64511789 | 0.29 |
ENST00000419843.1 ENST00000394430.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr12_-_51611477 | 0.29 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr1_-_156399184 | 0.29 |
ENST00000368243.1 ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr2_+_45878790 | 0.29 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr3_+_140660634 | 0.29 |
ENST00000446041.2 ENST00000507429.1 ENST00000324194.6 |
SLC25A36 |
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr2_-_10588630 | 0.29 |
ENST00000234111.4 |
ODC1 |
ornithine decarboxylase 1 |
| chr17_-_45266542 | 0.29 |
ENST00000531206.1 ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27 |
cell division cycle 27 |
| chrX_-_119445263 | 0.29 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 4.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 3.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 9.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 4.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 3.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 4.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 4.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.2 | 3.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 1.1 | 4.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.9 | 2.6 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.6 | 7.8 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.5 | 3.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.5 | 1.5 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 1.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 4.3 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.4 | 4.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.3 | 1.0 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.3 | 2.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.5 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 2.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.3 | 14.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.3 | 0.5 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.3 | 1.5 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 2.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 1.0 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 3.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.9 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.2 | 1.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 1.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 0.5 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.7 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.6 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.7 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.9 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.1 | 4.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.8 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.5 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 1.5 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.1 | 1.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.9 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.4 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 2.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.5 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.1 | 0.5 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 3.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 5.6 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.6 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.4 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.7 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 2.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:1901657 | glycosyl compound metabolic process(GO:1901657) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 1.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.4 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.3 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:1901569 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 2.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.3 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.1 | 4.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 3.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.4 | 4.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 4.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 4.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.3 | 0.8 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.3 | 1.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 4.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 2.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 3.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.4 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 0.9 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 8.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 2.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 16.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 2.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.4 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 4.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.0 | GO:0016020 | membrane(GO:0016020) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 2.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.4 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 1.4 | 4.3 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 1.4 | 4.2 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 1.0 | 3.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.4 | 1.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 2.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 1.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.7 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.2 | 3.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 0.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 2.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 0.5 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.2 | 4.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 1.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 3.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.7 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.2 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 3.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 1.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 3.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.0 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 3.2 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 2.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |