Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
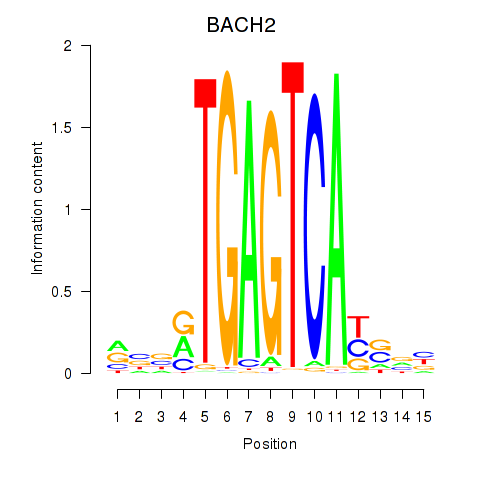
Results for BACH2
Z-value: 1.01
Transcription factors associated with BACH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH2
|
ENSG00000112182.10 | BACH2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BACH2 | hg19_v2_chr6_-_91006461_91006517, hg19_v2_chr6_-_91006627_91006641 | -0.45 | 7.7e-02 | Click! |
Activity profile of BACH2 motif
Sorted Z-values of BACH2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21452804 | 2.38 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr5_-_141061759 | 2.36 |
ENST00000508305.1 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr18_+_21452964 | 2.32 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr5_-_141061777 | 2.29 |
ENST00000239440.4 |
ARAP3 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr17_-_39769005 | 2.28 |
ENST00000301653.4 ENST00000593067.1 |
KRT16 |
keratin 16 |
| chr11_-_102668879 | 2.04 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chrX_-_154563889 | 1.72 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chrX_-_15511438 | 1.61 |
ENST00000380420.5 |
PIR |
pirin (iron-binding nuclear protein) |
| chr1_+_153003671 | 1.42 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr5_-_54281491 | 1.41 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr6_+_44194762 | 1.27 |
ENST00000371708.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr19_-_39264072 | 1.27 |
ENST00000599035.1 ENST00000378626.4 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
| chr1_+_155107820 | 1.20 |
ENST00000484157.1 |
SLC50A1 |
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr11_+_59522837 | 1.03 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr19_+_39279838 | 1.01 |
ENST00000314980.4 |
LGALS7B |
lectin, galactoside-binding, soluble, 7B |
| chr6_-_84140757 | 0.99 |
ENST00000541327.1 ENST00000369705.3 ENST00000543031.1 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr10_+_17270214 | 0.94 |
ENST00000544301.1 |
VIM |
vimentin |
| chr18_+_61143994 | 0.89 |
ENST00000382771.4 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr9_-_124991124 | 0.88 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr1_+_223889285 | 0.86 |
ENST00000433674.2 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr18_+_61442629 | 0.85 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr15_+_75335604 | 0.85 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr3_-_151034734 | 0.84 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr17_+_30771279 | 0.83 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr16_-_89043377 | 0.83 |
ENST00000436887.2 ENST00000448839.1 ENST00000360302.2 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr1_+_152881014 | 0.79 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr6_-_159239257 | 0.79 |
ENST00000337147.7 ENST00000392177.4 |
EZR |
ezrin |
| chr18_-_268019 | 0.77 |
ENST00000261600.6 |
THOC1 |
THO complex 1 |
| chr2_+_47596287 | 0.75 |
ENST00000263735.4 |
EPCAM |
epithelial cell adhesion molecule |
| chr6_-_35888905 | 0.75 |
ENST00000510290.1 ENST00000423325.2 ENST00000373822.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr15_+_44829334 | 0.71 |
ENST00000535391.1 |
EIF3J |
eukaryotic translation initiation factor 3, subunit J |
| chrX_+_12993202 | 0.69 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr1_-_158656488 | 0.67 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chrX_-_129244655 | 0.67 |
ENST00000335997.7 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
| chr17_-_73150629 | 0.67 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
| chr6_+_150070857 | 0.65 |
ENST00000544496.1 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr11_-_62457371 | 0.64 |
ENST00000317449.4 |
LRRN4CL |
LRRN4 C-terminal like |
| chr1_-_207224307 | 0.63 |
ENST00000315927.4 |
YOD1 |
YOD1 deubiquitinase |
| chr6_-_35888824 | 0.63 |
ENST00000361690.3 ENST00000512445.1 |
SRPK1 |
SRSF protein kinase 1 |
| chr12_-_48152853 | 0.62 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr2_-_85641162 | 0.61 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr17_+_7590734 | 0.60 |
ENST00000457584.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr1_+_36621697 | 0.60 |
ENST00000373150.4 ENST00000373151.2 |
MAP7D1 |
MAP7 domain containing 1 |
| chr4_+_74606223 | 0.60 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr1_+_36621529 | 0.60 |
ENST00000316156.4 |
MAP7D1 |
MAP7 domain containing 1 |
| chrX_-_129244454 | 0.60 |
ENST00000308167.5 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
| chr11_-_119993979 | 0.60 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_64013288 | 0.56 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr4_-_146019693 | 0.56 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr7_+_148395959 | 0.55 |
ENST00000325222.4 |
CUL1 |
cullin 1 |
| chr16_+_57662419 | 0.54 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57662138 | 0.54 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_-_64013663 | 0.53 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr10_-_6019984 | 0.53 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr14_-_23426337 | 0.53 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr12_-_102591604 | 0.53 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr12_-_48152611 | 0.52 |
ENST00000389212.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr4_+_170581213 | 0.52 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr14_-_23426322 | 0.51 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr10_-_6019552 | 0.50 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr17_-_48785216 | 0.50 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr16_+_58533951 | 0.50 |
ENST00000566192.1 ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4 |
NDRG family member 4 |
| chrX_-_16730984 | 0.50 |
ENST00000380241.3 |
CTPS2 |
CTP synthase 2 |
| chr14_-_23426270 | 0.49 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr11_+_10476851 | 0.48 |
ENST00000396553.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr6_+_292051 | 0.48 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr6_+_2988847 | 0.48 |
ENST00000380472.3 ENST00000605901.1 ENST00000454015.1 |
NQO2 LINC01011 |
NAD(P)H dehydrogenase, quinone 2 long intergenic non-protein coding RNA 1011 |
| chr4_-_100356844 | 0.47 |
ENST00000437033.2 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_+_150070831 | 0.47 |
ENST00000367380.5 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr7_+_99006232 | 0.46 |
ENST00000403633.2 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr22_+_23077065 | 0.46 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr15_+_89182178 | 0.46 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr3_+_19988885 | 0.45 |
ENST00000422242.1 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr19_-_51523412 | 0.45 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr1_-_179112173 | 0.44 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr6_+_32121218 | 0.44 |
ENST00000414204.1 ENST00000361568.2 ENST00000395523.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr3_+_11178779 | 0.44 |
ENST00000438284.2 |
HRH1 |
histamine receptor H1 |
| chr12_-_53343602 | 0.44 |
ENST00000546897.1 ENST00000552551.1 |
KRT8 |
keratin 8 |
| chr16_+_22308717 | 0.44 |
ENST00000299853.5 ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr4_-_100356291 | 0.43 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr13_+_32838801 | 0.43 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chrX_-_15288154 | 0.43 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chrX_-_16730688 | 0.42 |
ENST00000359276.4 |
CTPS2 |
CTP synthase 2 |
| chrX_+_41193407 | 0.42 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr20_+_44637526 | 0.42 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr10_+_104155450 | 0.42 |
ENST00000471698.1 ENST00000189444.6 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr1_-_179112189 | 0.40 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr9_+_4985016 | 0.40 |
ENST00000539801.1 |
JAK2 |
Janus kinase 2 |
| chr9_-_127177703 | 0.40 |
ENST00000259457.3 ENST00000536392.1 ENST00000441097.1 |
PSMB7 |
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr5_+_159436120 | 0.39 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr5_+_150020214 | 0.39 |
ENST00000307662.4 |
SYNPO |
synaptopodin |
| chr16_+_30968615 | 0.38 |
ENST00000262519.8 |
SETD1A |
SET domain containing 1A |
| chr1_-_27816641 | 0.38 |
ENST00000430629.2 |
WASF2 |
WAS protein family, member 2 |
| chr15_+_44829255 | 0.38 |
ENST00000261868.5 ENST00000424492.3 |
EIF3J |
eukaryotic translation initiation factor 3, subunit J |
| chr15_+_89182156 | 0.37 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr19_-_51523275 | 0.36 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr4_-_100356551 | 0.36 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_38670897 | 0.35 |
ENST00000373365.4 |
GLO1 |
glyoxalase I |
| chr5_+_110559784 | 0.35 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr17_-_42345487 | 0.34 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr2_+_169926047 | 0.33 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr12_-_719573 | 0.33 |
ENST00000397265.3 |
NINJ2 |
ninjurin 2 |
| chr17_+_30677136 | 0.32 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr22_+_23134974 | 0.32 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr12_-_71182695 | 0.32 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr22_+_22681656 | 0.31 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr5_+_110559603 | 0.31 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr15_+_89181974 | 0.30 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr9_+_4985228 | 0.30 |
ENST00000381652.3 |
JAK2 |
Janus kinase 2 |
| chr6_-_31704282 | 0.29 |
ENST00000375784.3 ENST00000375779.2 |
CLIC1 |
chloride intracellular channel 1 |
| chr12_-_7818474 | 0.29 |
ENST00000229304.4 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr5_-_175843524 | 0.28 |
ENST00000502877.1 |
CLTB |
clathrin, light chain B |
| chr7_+_100026406 | 0.28 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chr2_+_201980827 | 0.27 |
ENST00000309955.3 ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr15_-_60690163 | 0.27 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr2_+_231921574 | 0.26 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr12_-_8218997 | 0.26 |
ENST00000307637.4 |
C3AR1 |
complement component 3a receptor 1 |
| chr7_+_123488124 | 0.26 |
ENST00000476325.1 |
HYAL4 |
hyaluronoglucosaminidase 4 |
| chr6_+_44215603 | 0.26 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr3_-_98241358 | 0.25 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr17_-_65362678 | 0.25 |
ENST00000357146.4 ENST00000356126.3 |
PSMD12 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr9_-_32573130 | 0.24 |
ENST00000350021.2 ENST00000379847.3 |
NDUFB6 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr14_+_103801140 | 0.23 |
ENST00000561325.1 ENST00000392715.2 ENST00000559130.1 ENST00000559532.1 ENST00000558506.1 |
EIF5 |
eukaryotic translation initiation factor 5 |
| chr10_-_23003460 | 0.23 |
ENST00000376573.4 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr3_-_56502375 | 0.23 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr7_+_150065278 | 0.22 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr5_-_175843569 | 0.22 |
ENST00000310418.4 ENST00000345807.2 |
CLTB |
clathrin, light chain B |
| chr6_+_44214824 | 0.21 |
ENST00000371646.5 ENST00000353801.3 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr22_+_22676808 | 0.21 |
ENST00000390290.2 |
IGLV1-51 |
immunoglobulin lambda variable 1-51 |
| chr10_+_121410882 | 0.20 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr17_+_55162453 | 0.20 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr1_+_223889310 | 0.19 |
ENST00000434648.1 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr11_+_46316677 | 0.19 |
ENST00000534787.1 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr6_+_106959718 | 0.19 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chr3_+_48507621 | 0.18 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr6_+_31082603 | 0.17 |
ENST00000259881.9 |
PSORS1C1 |
psoriasis susceptibility 1 candidate 1 |
| chr6_+_41604747 | 0.17 |
ENST00000419164.1 ENST00000373051.2 |
MDFI |
MyoD family inhibitor |
| chr22_+_22707260 | 0.16 |
ENST00000390293.1 |
IGLV5-48 |
immunoglobulin lambda variable 5-48 (non-functional) |
| chr5_-_75919253 | 0.16 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr3_-_49823941 | 0.15 |
ENST00000321599.4 ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1 |
inositol hexakisphosphate kinase 1 |
| chr16_+_2083265 | 0.15 |
ENST00000565855.1 ENST00000566198.1 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr22_+_22712087 | 0.15 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chr11_+_20620946 | 0.15 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr6_-_31745085 | 0.15 |
ENST00000375686.3 ENST00000447450.1 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr1_-_204121013 | 0.15 |
ENST00000367201.3 |
ETNK2 |
ethanolamine kinase 2 |
| chr20_+_44034804 | 0.14 |
ENST00000357275.2 ENST00000372720.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_-_65593784 | 0.14 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr12_+_56732658 | 0.14 |
ENST00000228534.4 |
IL23A |
interleukin 23, alpha subunit p19 |
| chr19_-_51538148 | 0.14 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr10_+_88854926 | 0.13 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr9_+_140135665 | 0.12 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr19_-_51538118 | 0.12 |
ENST00000529888.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr10_+_88428370 | 0.12 |
ENST00000372066.3 ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3 |
LIM domain binding 3 |
| chr8_-_59572093 | 0.12 |
ENST00000427130.2 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr19_+_13842559 | 0.11 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr19_+_47835404 | 0.11 |
ENST00000595464.1 |
C5AR2 |
complement component 5a receptor 2 |
| chr17_-_59668550 | 0.11 |
ENST00000521764.1 |
NACA2 |
nascent polypeptide-associated complex alpha subunit 2 |
| chr20_-_17539456 | 0.11 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr22_+_22764088 | 0.11 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr22_+_23063100 | 0.10 |
ENST00000390309.2 |
IGLV3-19 |
immunoglobulin lambda variable 3-19 |
| chr6_-_31745037 | 0.10 |
ENST00000375688.4 |
VWA7 |
von Willebrand factor A domain containing 7 |
| chr1_-_204121102 | 0.10 |
ENST00000367202.4 |
ETNK2 |
ethanolamine kinase 2 |
| chr16_-_1429627 | 0.10 |
ENST00000248104.7 |
UNKL |
unkempt family zinc finger-like |
| chr5_-_75919217 | 0.10 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr10_+_85899196 | 0.10 |
ENST00000372134.3 |
GHITM |
growth hormone inducible transmembrane protein |
| chr1_-_204121298 | 0.10 |
ENST00000367199.2 |
ETNK2 |
ethanolamine kinase 2 |
| chr1_+_223900034 | 0.10 |
ENST00000295006.5 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr17_+_25799008 | 0.09 |
ENST00000583370.1 ENST00000398988.3 ENST00000268763.6 |
KSR1 |
kinase suppressor of ras 1 |
| chr17_-_18161870 | 0.09 |
ENST00000579294.1 ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII |
flightless I homolog (Drosophila) |
| chr19_-_48018203 | 0.09 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr4_-_74088800 | 0.09 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr11_+_10477733 | 0.09 |
ENST00000528723.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr1_-_109968973 | 0.08 |
ENST00000271308.4 ENST00000538610.1 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr14_-_25103388 | 0.08 |
ENST00000526004.1 ENST00000415355.3 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_-_25103472 | 0.08 |
ENST00000216341.4 ENST00000382542.1 ENST00000382540.1 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_-_11610255 | 0.08 |
ENST00000424529.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr5_+_72143988 | 0.07 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chr3_+_48507210 | 0.07 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr2_-_220173685 | 0.07 |
ENST00000423636.2 ENST00000442029.1 ENST00000412847.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr22_+_22735135 | 0.07 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr2_-_11606275 | 0.06 |
ENST00000381525.3 ENST00000362009.4 |
E2F6 |
E2F transcription factor 6 |
| chr7_-_135433460 | 0.06 |
ENST00000415751.1 |
FAM180A |
family with sequence similarity 180, member A |
| chr2_-_89459813 | 0.06 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr12_-_76478686 | 0.06 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr1_-_51796987 | 0.06 |
ENST00000262676.5 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr7_-_96339132 | 0.06 |
ENST00000413065.1 |
SHFM1 |
split hand/foot malformation (ectrodactyly) type 1 |
| chr9_-_91793675 | 0.05 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr12_+_64845660 | 0.05 |
ENST00000331710.5 |
TBK1 |
TANK-binding kinase 1 |
| chr1_-_11918988 | 0.05 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr7_+_30960915 | 0.05 |
ENST00000441328.2 ENST00000409899.1 ENST00000409611.1 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr18_+_268148 | 0.05 |
ENST00000581677.1 |
RP11-705O1.8 |
RP11-705O1.8 |
| chr16_-_1429674 | 0.05 |
ENST00000403703.1 ENST00000397464.1 ENST00000402641.2 |
UNKL |
unkempt family zinc finger-like |
| chr2_+_233562015 | 0.05 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr11_+_5009424 | 0.05 |
ENST00000300762.1 |
MMP26 |
matrix metallopeptidase 26 |
| chr22_+_20861858 | 0.05 |
ENST00000414658.1 ENST00000432052.1 ENST00000425759.2 ENST00000292733.7 ENST00000542773.1 ENST00000263205.7 ENST00000406969.1 ENST00000382974.2 |
MED15 |
mediator complex subunit 15 |
| chr17_-_73775839 | 0.04 |
ENST00000592643.1 ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B |
H3 histone, family 3B (H3.3B) |
| chr1_+_19638788 | 0.04 |
ENST00000375155.3 ENST00000375153.3 ENST00000400548.2 |
PQLC2 |
PQ loop repeat containing 2 |
| chr7_-_98467629 | 0.04 |
ENST00000339375.4 |
TMEM130 |
transmembrane protein 130 |
| chr9_-_114246635 | 0.04 |
ENST00000338205.5 |
KIAA0368 |
KIAA0368 |
| chr1_+_26606608 | 0.04 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 4.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.4 | 1.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.4 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 1.3 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.3 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 0.6 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.2 | 4.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 1.0 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.2 | 0.5 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 0.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.5 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 1.0 | GO:0050544 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.2 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 2.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0043495 | sodium:bicarbonate symporter activity(GO:0008510) protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 1.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 1.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.4 | 1.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.4 | 1.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 1.1 | GO:0030822 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.3 | 0.8 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.3 | 1.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 0.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.7 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.2 | 0.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 0.6 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.2 | 2.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.2 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.1 | 1.0 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.4 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 1.4 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.7 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 0.5 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.7 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.4 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.3 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 | 0.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.4 | GO:0071651 | mature ribosome assembly(GO:0042256) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.5 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.1 | 0.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.7 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.1 | 0.5 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.0 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.7 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.8 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:1903401 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 1.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition(GO:0051437) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.0 | 0.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 0.8 | GO:0044393 | microspike(GO:0044393) |
| 0.3 | 1.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |