Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
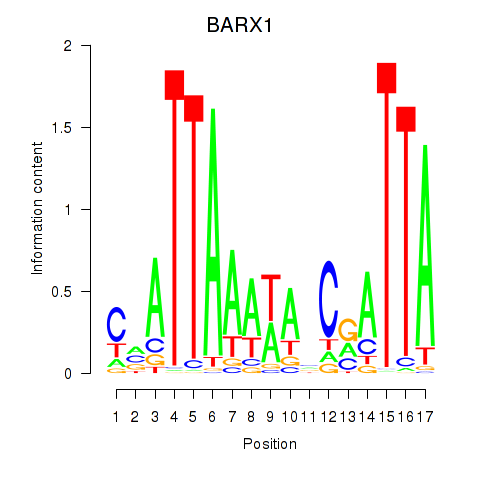
Results for BARX1
Z-value: 1.09
Transcription factors associated with BARX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARX1
|
ENSG00000131668.9 | BARX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BARX1 | hg19_v2_chr9_-_96717654_96717666 | 0.16 | 5.5e-01 | Click! |
Activity profile of BARX1 motif
Sorted Z-values of BARX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BARX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52911718 | 3.77 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr10_-_95242044 | 2.80 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr10_-_95241951 | 2.77 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr13_-_38172863 | 2.45 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr4_+_169418195 | 2.41 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr18_+_61442629 | 2.39 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr11_+_12766583 | 2.35 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr8_+_30244580 | 2.21 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr7_+_134430212 | 2.18 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr1_-_95391315 | 2.03 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr3_-_149093499 | 1.98 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr5_+_150404904 | 1.78 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr4_+_169418255 | 1.74 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr2_-_106054952 | 1.73 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr12_-_8803128 | 1.64 |
ENST00000543467.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr13_+_73629107 | 1.56 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr9_-_113761720 | 1.53 |
ENST00000541779.1 ENST00000374430.2 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr2_-_175711133 | 1.53 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr3_-_185538849 | 1.53 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr11_+_114168085 | 1.51 |
ENST00000541754.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr12_+_26348246 | 1.48 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr12_-_91572278 | 1.46 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr11_+_35201826 | 1.43 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chrX_-_13835147 | 1.42 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr5_+_140762268 | 1.41 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr7_+_32996997 | 1.41 |
ENST00000242209.4 ENST00000538336.1 ENST00000538443.1 |
FKBP9 |
FK506 binding protein 9, 63 kDa |
| chr14_+_55595762 | 1.40 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr22_-_36220420 | 1.37 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_123512688 | 1.35 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr18_+_21529811 | 1.26 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr5_-_16742330 | 1.22 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr4_-_7873981 | 1.20 |
ENST00000360265.4 |
AFAP1 |
actin filament associated protein 1 |
| chr12_-_91574142 | 1.20 |
ENST00000547937.1 |
DCN |
decorin |
| chr6_-_52859046 | 1.19 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr10_-_91011548 | 1.14 |
ENST00000456827.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr16_-_10652993 | 1.13 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr7_+_134576317 | 1.12 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr12_-_91546926 | 1.11 |
ENST00000550758.1 |
DCN |
decorin |
| chr1_+_73771844 | 1.07 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr10_-_79398250 | 1.07 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_44117260 | 1.07 |
ENST00000358681.4 |
EXT2 |
exostosin glycosyltransferase 2 |
| chrX_-_114252193 | 1.05 |
ENST00000243213.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chrX_-_114253536 | 1.03 |
ENST00000371936.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chr15_-_56757329 | 0.99 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr7_+_134576151 | 0.99 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr11_+_44117099 | 0.95 |
ENST00000533608.1 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr12_+_26274917 | 0.94 |
ENST00000538142.1 |
SSPN |
sarcospan |
| chr9_+_139874683 | 0.94 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr1_+_196788887 | 0.94 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr4_+_26585538 | 0.93 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr21_+_39644305 | 0.89 |
ENST00000398930.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_31361543 | 0.89 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr4_+_26585686 | 0.88 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr1_-_227505289 | 0.86 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr14_+_55595960 | 0.85 |
ENST00000554715.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr12_+_19282643 | 0.85 |
ENST00000317589.4 ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr17_+_18601299 | 0.85 |
ENST00000572555.1 ENST00000395902.3 ENST00000449552.2 |
TRIM16L |
tripartite motif containing 16-like |
| chr6_-_2971429 | 0.84 |
ENST00000380529.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chrX_-_41449204 | 0.84 |
ENST00000378179.3 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr17_+_61086917 | 0.83 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr1_+_78470530 | 0.82 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr19_-_55660561 | 0.82 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr3_-_151034734 | 0.82 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr8_+_9046503 | 0.82 |
ENST00000512942.2 |
RP11-10A14.5 |
RP11-10A14.5 |
| chr5_-_125930929 | 0.80 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr19_+_41281416 | 0.78 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr9_-_21974820 | 0.76 |
ENST00000579122.1 ENST00000498124.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr5_+_140743859 | 0.76 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr2_+_234621551 | 0.75 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_-_190927447 | 0.75 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr19_-_36643329 | 0.75 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr13_+_102142296 | 0.73 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_10978957 | 0.71 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr7_+_16793160 | 0.71 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr1_+_156030937 | 0.66 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chrX_-_134156502 | 0.66 |
ENST00000391440.1 |
FAM127C |
family with sequence similarity 127, member C |
| chr1_+_172389821 | 0.64 |
ENST00000367727.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_-_109584768 | 0.64 |
ENST00000357672.3 |
WDR47 |
WD repeat domain 47 |
| chr16_-_66584059 | 0.64 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr16_+_14802801 | 0.63 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr7_+_93551011 | 0.63 |
ENST00000248564.5 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
| chr18_+_61254534 | 0.63 |
ENST00000269489.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr7_+_107220660 | 0.61 |
ENST00000465919.1 ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr20_+_11898507 | 0.60 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr10_-_79397740 | 0.59 |
ENST00000372440.1 ENST00000480683.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_-_43424500 | 0.58 |
ENST00000415851.2 ENST00000426263.3 ENST00000372500.3 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr16_-_29517141 | 0.58 |
ENST00000550665.1 |
RP11-231C14.4 |
Uncharacterized protein |
| chr14_+_22458631 | 0.57 |
ENST00000390444.1 |
TRAV16 |
T cell receptor alpha variable 16 |
| chr3_+_138066539 | 0.56 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr4_-_57524061 | 0.56 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr9_-_95244781 | 0.56 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr2_+_130737223 | 0.55 |
ENST00000410061.2 |
RAB6C |
RAB6C, member RAS oncogene family |
| chr1_-_21377447 | 0.55 |
ENST00000374937.3 ENST00000264211.8 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_+_53659746 | 0.55 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr7_+_18535786 | 0.54 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr3_+_8543393 | 0.53 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_210517895 | 0.52 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr12_+_26348429 | 0.52 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr9_-_21351377 | 0.52 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr1_-_146082633 | 0.49 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr7_-_137028498 | 0.49 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr9_-_13279563 | 0.48 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr20_-_43150601 | 0.48 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr1_-_149908217 | 0.47 |
ENST00000369140.3 |
MTMR11 |
myotubularin related protein 11 |
| chr2_+_109237717 | 0.47 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr16_-_18462221 | 0.46 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr1_-_6420737 | 0.45 |
ENST00000541130.1 ENST00000377845.3 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr12_+_104609550 | 0.45 |
ENST00000525566.1 ENST00000429002.2 |
TXNRD1 |
thioredoxin reductase 1 |
| chr15_+_71228826 | 0.45 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr5_+_140797296 | 0.45 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr11_-_118134997 | 0.43 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr12_-_27167233 | 0.42 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
| chr2_+_172544294 | 0.42 |
ENST00000358002.6 ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr5_+_140734570 | 0.41 |
ENST00000571252.1 |
PCDHGA4 |
protocadherin gamma subfamily A, 4 |
| chr15_+_72978539 | 0.41 |
ENST00000539603.1 ENST00000569338.1 |
BBS4 |
Bardet-Biedl syndrome 4 |
| chr14_-_75643296 | 0.41 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr7_+_107220422 | 0.40 |
ENST00000005259.4 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr1_-_21377383 | 0.40 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_-_74618964 | 0.40 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr12_+_110011571 | 0.40 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr16_+_28648975 | 0.40 |
ENST00000529716.1 |
NPIPB8 |
nuclear pore complex interacting protein family, member B8 |
| chr5_+_147443534 | 0.39 |
ENST00000398454.1 ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5 |
serine peptidase inhibitor, Kazal type 5 |
| chr16_+_22501658 | 0.39 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr15_+_72978521 | 0.39 |
ENST00000542334.1 ENST00000268057.4 |
BBS4 |
Bardet-Biedl syndrome 4 |
| chr5_+_140792614 | 0.39 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr18_-_61311485 | 0.38 |
ENST00000436264.1 ENST00000356424.6 ENST00000341074.5 |
SERPINB4 |
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr10_-_79398127 | 0.38 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_11062161 | 0.38 |
ENST00000390677.2 |
TAS2R13 |
taste receptor, type 2, member 13 |
| chr1_+_171283331 | 0.37 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chr4_+_177241094 | 0.37 |
ENST00000503362.1 |
SPCS3 |
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr22_-_19466732 | 0.36 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr17_-_41174424 | 0.35 |
ENST00000355653.3 |
VAT1 |
vesicle amine transport 1 |
| chr12_-_10151773 | 0.35 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr17_-_71410794 | 0.35 |
ENST00000424778.1 |
SDK2 |
sidekick cell adhesion molecule 2 |
| chr22_+_45148432 | 0.34 |
ENST00000389774.2 ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8 |
Rho GTPase activating protein 8 |
| chr1_-_165668100 | 0.34 |
ENST00000354775.4 |
ALDH9A1 |
aldehyde dehydrogenase 9 family, member A1 |
| chr1_+_84630645 | 0.34 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_-_138763734 | 0.34 |
ENST00000413199.1 ENST00000502927.2 |
PRR23C |
proline rich 23C |
| chr8_+_54764346 | 0.34 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr11_-_86383650 | 0.33 |
ENST00000526944.1 ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr4_+_492985 | 0.33 |
ENST00000296306.7 ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG |
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr1_+_87012753 | 0.33 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr13_-_33780133 | 0.33 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chrX_-_104465358 | 0.33 |
ENST00000372578.3 ENST00000372575.1 ENST00000413579.1 |
TEX13A |
testis expressed 13A |
| chr1_-_246729544 | 0.31 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr22_-_38669030 | 0.31 |
ENST00000361906.3 |
TMEM184B |
transmembrane protein 184B |
| chr12_+_123237321 | 0.31 |
ENST00000280557.6 ENST00000455982.2 |
DENR |
density-regulated protein |
| chr1_-_153085984 | 0.31 |
ENST00000468739.1 |
SPRR2F |
small proline-rich protein 2F |
| chr17_-_19266045 | 0.31 |
ENST00000395616.3 |
B9D1 |
B9 protein domain 1 |
| chr19_-_47349395 | 0.31 |
ENST00000597020.1 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_-_20251744 | 0.31 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr10_+_70748487 | 0.31 |
ENST00000361983.4 |
KIAA1279 |
KIAA1279 |
| chr17_-_7145475 | 0.31 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr4_-_69536346 | 0.31 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr21_-_35884573 | 0.31 |
ENST00000399286.2 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
| chr1_-_203151933 | 0.31 |
ENST00000404436.2 |
CHI3L1 |
chitinase 3-like 1 (cartilage glycoprotein-39) |
| chr1_-_153029980 | 0.31 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr13_+_77564795 | 0.30 |
ENST00000377453.3 |
CLN5 |
ceroid-lipofuscinosis, neuronal 5 |
| chr5_-_9630463 | 0.30 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr1_+_196912902 | 0.30 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr5_-_55412774 | 0.30 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr3_-_151047327 | 0.29 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr7_-_72993033 | 0.29 |
ENST00000305632.5 |
TBL2 |
transducin (beta)-like 2 |
| chr5_+_140552218 | 0.29 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr13_+_48611665 | 0.28 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr4_+_169013666 | 0.28 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr12_-_10573149 | 0.28 |
ENST00000381904.2 ENST00000381903.2 ENST00000396439.2 |
KLRC3 |
killer cell lectin-like receptor subfamily C, member 3 |
| chr22_-_19466683 | 0.28 |
ENST00000399523.1 ENST00000421968.2 ENST00000447868.1 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr3_+_189349162 | 0.28 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr9_+_35161998 | 0.27 |
ENST00000396787.1 ENST00000378495.3 ENST00000378496.4 |
UNC13B |
unc-13 homolog B (C. elegans) |
| chr5_+_140474181 | 0.27 |
ENST00000194155.4 |
PCDHB2 |
protocadherin beta 2 |
| chr22_+_51176624 | 0.27 |
ENST00000216139.5 ENST00000529621.1 |
ACR |
acrosin |
| chr15_-_72523924 | 0.26 |
ENST00000566809.1 ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM |
pyruvate kinase, muscle |
| chr12_-_10962767 | 0.26 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr12_+_122326662 | 0.25 |
ENST00000261817.2 ENST00000538613.1 ENST00000542602.1 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr21_+_47531328 | 0.25 |
ENST00000409416.1 ENST00000397763.1 |
COL6A2 |
collagen, type VI, alpha 2 |
| chr7_+_106505696 | 0.25 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr4_-_57547454 | 0.25 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr17_-_66951474 | 0.25 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr4_+_95376396 | 0.25 |
ENST00000508216.1 ENST00000514743.1 |
PDLIM5 |
PDZ and LIM domain 5 |
| chr17_-_3301704 | 0.25 |
ENST00000322608.2 |
OR1E1 |
olfactory receptor, family 1, subfamily E, member 1 |
| chr8_-_141774467 | 0.24 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr16_-_28374829 | 0.24 |
ENST00000532254.1 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
| chr2_-_74619152 | 0.24 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr4_-_70518941 | 0.24 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr18_+_66465302 | 0.24 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr15_-_78913521 | 0.24 |
ENST00000326828.5 |
CHRNA3 |
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr10_-_92681033 | 0.24 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr12_-_71551652 | 0.23 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr20_+_123010 | 0.23 |
ENST00000382398.3 |
DEFB126 |
defensin, beta 126 |
| chr5_-_93447333 | 0.23 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr3_+_8543533 | 0.23 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_90259593 | 0.23 |
ENST00000471857.1 |
IGKV1D-8 |
immunoglobulin kappa variable 1D-8 |
| chr7_-_105752971 | 0.23 |
ENST00000011473.2 |
SYPL1 |
synaptophysin-like 1 |
| chr19_+_9361606 | 0.23 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr20_-_10414804 | 0.23 |
ENST00000347364.3 |
MKKS |
McKusick-Kaufman syndrome |
| chr17_-_2966901 | 0.23 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr6_-_136788001 | 0.23 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr6_-_52705641 | 0.22 |
ENST00000370989.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr15_-_89089860 | 0.22 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.7 | 2.2 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.6 | 2.4 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.6 | 1.8 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.5 | 5.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 1.7 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 1.0 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 3.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.5 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.2 | 2.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 1.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 2.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.6 | GO:0070837 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.8 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 2.0 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.6 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 4.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 5.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.6 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.5 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.8 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 1.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.1 | 0.4 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 1.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.8 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.5 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.4 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 1.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 2.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.7 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.8 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.3 | GO:0010482 | ectoderm and mesoderm interaction(GO:0007499) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 1.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.5 | GO:0070933 | peptidyl-lysine deacetylation(GO:0034983) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.3 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.6 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 1.1 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.2 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 0.2 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.7 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 1.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 1.2 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 1.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.2 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 4.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 1.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.7 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.8 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 6.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 2.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 4.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 3.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 8.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 1.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 2.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.0 | GO:0031673 | H zone(GO:0031673) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 5.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 5.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 5.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 1.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.3 | 2.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 2.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 2.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.3 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 1.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.6 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.2 | 0.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 5.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 4.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 1.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 1.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 2.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 2.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.4 | GO:0008948 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.1 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 1.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 3.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 4.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 3.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 1.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0019237 | satellite DNA binding(GO:0003696) centromeric DNA binding(GO:0019237) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |