Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
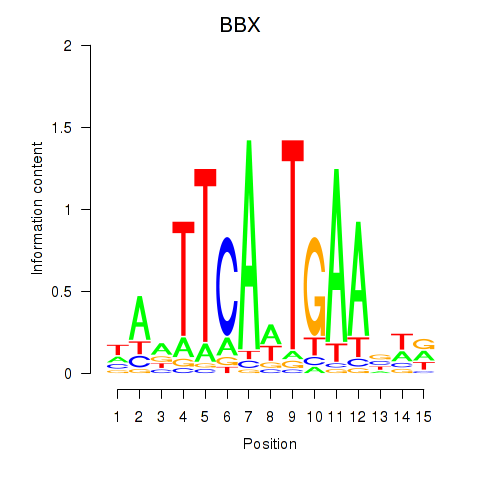
Results for BBX
Z-value: 0.62
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg19_v2_chr3_+_107241783_107241850 | 0.37 | 1.5e-01 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484155 | 2.34 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr9_-_116837249 | 2.11 |
ENST00000466610.2 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr4_+_155484103 | 1.80 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr4_-_70080449 | 1.50 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr9_-_5833027 | 1.17 |
ENST00000339450.5 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
| chr4_-_186697044 | 0.96 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_+_31895254 | 0.95 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr11_+_22689648 | 0.93 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr4_-_186733363 | 0.88 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_186696425 | 0.75 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr12_-_52887034 | 0.72 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr22_+_44319619 | 0.65 |
ENST00000216180.3 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr16_+_56672571 | 0.61 |
ENST00000290705.8 |
MT1A |
metallothionein 1A |
| chr13_+_50570019 | 0.60 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr1_-_59043166 | 0.37 |
ENST00000371225.2 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
| chr14_+_23235886 | 0.37 |
ENST00000604262.1 ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
| chr1_+_229440129 | 0.35 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr16_+_28763108 | 0.34 |
ENST00000357796.3 ENST00000550983.1 |
NPIPB9 |
nuclear pore complex interacting protein family, member B9 |
| chr5_-_78809950 | 0.32 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr1_+_55464600 | 0.30 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr2_+_101437487 | 0.30 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr17_+_66255310 | 0.27 |
ENST00000448504.2 |
ARSG |
arylsulfatase G |
| chr20_-_30310656 | 0.26 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr16_-_28374829 | 0.26 |
ENST00000532254.1 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
| chr5_+_140514782 | 0.25 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr20_-_56286479 | 0.24 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr5_+_140480083 | 0.24 |
ENST00000231130.2 |
PCDHB3 |
protocadherin beta 3 |
| chrX_+_54834004 | 0.24 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr9_+_35161998 | 0.23 |
ENST00000396787.1 ENST00000378495.3 ENST00000378496.4 |
UNC13B |
unc-13 homolog B (C. elegans) |
| chr14_+_94492674 | 0.20 |
ENST00000203664.5 ENST00000553723.1 |
OTUB2 |
OTU domain, ubiquitin aldehyde binding 2 |
| chr6_-_25874440 | 0.19 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr7_-_16872932 | 0.19 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr20_-_30310693 | 0.19 |
ENST00000307677.4 ENST00000420653.1 |
BCL2L1 |
BCL2-like 1 |
| chr19_+_47421933 | 0.19 |
ENST00000404338.3 |
ARHGAP35 |
Rho GTPase activating protein 35 |
| chr18_-_21017817 | 0.18 |
ENST00000542162.1 ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241 |
transmembrane protein 241 |
| chr2_-_62081254 | 0.17 |
ENST00000405894.3 |
FAM161A |
family with sequence similarity 161, member A |
| chr20_-_30310336 | 0.16 |
ENST00000434194.1 ENST00000376062.2 |
BCL2L1 |
BCL2-like 1 |
| chr2_-_62081148 | 0.15 |
ENST00000404929.1 |
FAM161A |
family with sequence similarity 161, member A |
| chr1_-_36948879 | 0.14 |
ENST00000373106.1 ENST00000373104.1 ENST00000373103.1 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr1_+_117963209 | 0.14 |
ENST00000449370.2 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
| chr4_+_69313145 | 0.13 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr6_-_150067696 | 0.13 |
ENST00000340413.2 ENST00000367403.3 |
NUP43 |
nucleoporin 43kDa |
| chr15_+_65823092 | 0.13 |
ENST00000566074.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr6_+_72922590 | 0.12 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_-_94003003 | 0.11 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr22_+_32871224 | 0.11 |
ENST00000452138.1 ENST00000382058.3 ENST00000397426.1 |
FBXO7 |
F-box protein 7 |
| chr11_+_118443098 | 0.11 |
ENST00000392859.3 ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1 |
archain 1 |
| chr6_+_72922505 | 0.11 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr12_-_10978957 | 0.10 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr13_+_33160553 | 0.10 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_+_148583043 | 0.09 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr13_+_28494130 | 0.08 |
ENST00000381033.4 |
PDX1 |
pancreatic and duodenal homeobox 1 |
| chr8_-_6783588 | 0.07 |
ENST00000297436.2 |
DEFA6 |
defensin, alpha 6, Paneth cell-specific |
| chr2_-_55237484 | 0.07 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr1_+_52082751 | 0.07 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr1_-_216978709 | 0.06 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chrX_-_140786896 | 0.04 |
ENST00000370515.3 |
SPANXD |
SPANX family, member D |
| chr12_-_21487829 | 0.03 |
ENST00000445053.1 ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2 |
solute carrier organic anion transporter family, member 1A2 |
| chrX_+_140677562 | 0.03 |
ENST00000370518.3 |
SPANXA2 |
SPANX family, member A2 |
| chr17_-_8263538 | 0.02 |
ENST00000535173.1 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr14_+_22337014 | 0.02 |
ENST00000390436.2 |
TRAV13-1 |
T cell receptor alpha variable 13-1 |
| chr11_+_58390132 | 0.02 |
ENST00000361987.4 |
CNTF |
ciliary neurotrophic factor |
| chr19_+_7828035 | 0.02 |
ENST00000327325.5 ENST00000394122.2 ENST00000248228.4 ENST00000334806.5 ENST00000359059.5 ENST00000357361.2 ENST00000596363.1 ENST00000595751.1 ENST00000596707.1 ENST00000597522.1 ENST00000595496.1 |
CLEC4M |
C-type lectin domain family 4, member M |
| chr3_+_111260856 | 0.01 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr17_+_45331184 | 0.01 |
ENST00000559488.1 ENST00000571680.1 ENST00000435993.2 |
ITGB3 |
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 4.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.7 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 2.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.4 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.6 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 2.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.6 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 1.5 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |