Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for BCL6B
Z-value: 0.80
Transcription factors associated with BCL6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BCL6B
|
ENSG00000161940.6 | BCL6B |
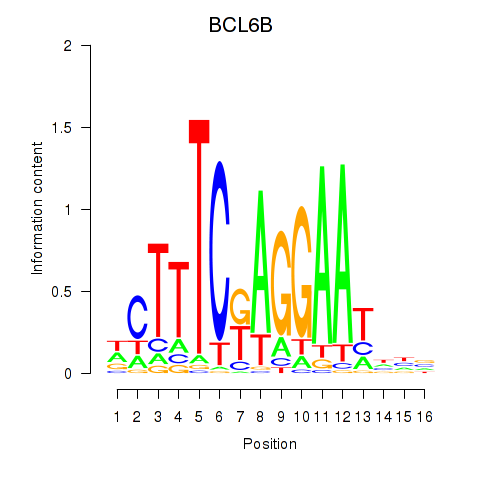
Activity profile of BCL6B motif
Sorted Z-values of BCL6B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BCL6B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123512688 | 3.38 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr12_-_91574142 | 3.00 |
ENST00000547937.1 |
DCN |
decorin |
| chr2_-_216257849 | 2.75 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr2_-_188419200 | 2.59 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_188419078 | 2.23 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr4_-_99578789 | 2.01 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr7_+_134464414 | 1.86 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr3_-_123603137 | 1.85 |
ENST00000360304.3 ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK |
myosin light chain kinase |
| chr4_-_99578776 | 1.75 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr12_+_19358228 | 1.70 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr6_+_7541845 | 1.66 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr14_+_95078714 | 1.53 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr7_+_134464376 | 1.48 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr6_+_7541808 | 1.38 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr1_-_108735440 | 1.17 |
ENST00000370041.4 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr1_-_31845914 | 1.09 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr16_-_18441131 | 1.09 |
ENST00000339303.5 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr9_-_117880477 | 1.01 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr14_-_30396948 | 0.86 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr1_-_230850043 | 0.85 |
ENST00000366667.4 |
AGT |
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chrX_-_153775426 | 0.83 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr16_+_16481598 | 0.80 |
ENST00000327792.5 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr5_-_172198190 | 0.80 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr7_+_872107 | 0.75 |
ENST00000405266.1 ENST00000401592.1 ENST00000403868.1 ENST00000425407.2 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
| chr2_-_55237484 | 0.74 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr3_+_158787041 | 0.73 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr8_-_141810634 | 0.66 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr5_-_140700322 | 0.55 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr1_+_84609944 | 0.55 |
ENST00000370685.3 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_155511887 | 0.52 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr10_-_70287231 | 0.49 |
ENST00000609923.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr11_-_18258342 | 0.49 |
ENST00000278222.4 |
SAA4 |
serum amyloid A4, constitutive |
| chr17_-_42907564 | 0.48 |
ENST00000592524.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chr17_-_48474828 | 0.48 |
ENST00000576448.1 ENST00000225972.7 |
LRRC59 |
leucine rich repeat containing 59 |
| chr19_-_44860820 | 0.45 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr6_+_31588478 | 0.41 |
ENST00000376007.4 ENST00000376033.2 |
PRRC2A |
proline-rich coiled-coil 2A |
| chr7_-_29186008 | 0.38 |
ENST00000396276.3 ENST00000265394.5 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr1_+_221051699 | 0.36 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr11_+_73358594 | 0.36 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr19_+_45147098 | 0.34 |
ENST00000425690.3 ENST00000344956.4 ENST00000403059.4 |
PVR |
poliovirus receptor |
| chr13_-_33112823 | 0.34 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr19_+_45147313 | 0.31 |
ENST00000406449.4 |
PVR |
poliovirus receptor |
| chr4_+_86396265 | 0.31 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr11_+_123986069 | 0.31 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr15_+_65843130 | 0.30 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr10_-_70287172 | 0.29 |
ENST00000539557.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr20_+_56136136 | 0.29 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr8_+_104427581 | 0.29 |
ENST00000521716.1 ENST00000521971.1 ENST00000519682.1 |
DCAF13 |
DDB1 and CUL4 associated factor 13 |
| chr17_-_42908155 | 0.28 |
ENST00000426548.1 ENST00000590758.1 ENST00000591424.1 |
GJC1 |
gap junction protein, gamma 1, 45kDa |
| chrX_+_153775821 | 0.27 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr19_+_11200038 | 0.27 |
ENST00000558518.1 ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR |
low density lipoprotein receptor |
| chr13_-_36705425 | 0.26 |
ENST00000255448.4 ENST00000360631.3 ENST00000379892.4 |
DCLK1 |
doublecortin-like kinase 1 |
| chr11_+_27062860 | 0.23 |
ENST00000528583.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_+_39760129 | 0.23 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr12_-_120687948 | 0.22 |
ENST00000458477.2 |
PXN |
paxillin |
| chr1_-_150669604 | 0.22 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr12_+_104324112 | 0.22 |
ENST00000299767.5 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr5_+_140480083 | 0.20 |
ENST00000231130.2 |
PCDHB3 |
protocadherin beta 3 |
| chr12_+_16064106 | 0.20 |
ENST00000428559.2 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chrX_+_95939638 | 0.19 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chrX_-_135338503 | 0.19 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chr12_-_10151773 | 0.19 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr3_+_46618727 | 0.19 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr15_+_45003675 | 0.17 |
ENST00000558401.1 ENST00000559916.1 ENST00000544417.1 |
B2M |
beta-2-microglobulin |
| chr5_+_54398463 | 0.16 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr2_-_31361543 | 0.16 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr6_+_125540951 | 0.16 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr4_+_70796784 | 0.16 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr12_+_93964746 | 0.13 |
ENST00000536696.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr19_-_7812397 | 0.13 |
ENST00000593660.1 ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209 |
CD209 molecule |
| chr1_+_235530675 | 0.12 |
ENST00000366601.3 ENST00000406207.1 ENST00000543662.1 |
TBCE |
tubulin folding cofactor E |
| chr12_+_8975061 | 0.12 |
ENST00000299698.7 |
A2ML1 |
alpha-2-macroglobulin-like 1 |
| chr1_-_8000872 | 0.12 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr1_+_209878182 | 0.11 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr5_+_140579162 | 0.10 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr5_+_80529104 | 0.09 |
ENST00000254035.4 ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
| chrX_-_57147748 | 0.09 |
ENST00000374910.3 |
SPIN2B |
spindlin family, member 2B |
| chr6_-_87804815 | 0.09 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr6_-_155776966 | 0.09 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr5_+_140501581 | 0.08 |
ENST00000194152.1 |
PCDHB4 |
protocadherin beta 4 |
| chr12_-_11002063 | 0.07 |
ENST00000544994.1 ENST00000228811.4 ENST00000540107.1 |
PRR4 |
proline rich 4 (lacrimal) |
| chr5_+_140535577 | 0.07 |
ENST00000539533.1 |
PCDHB17 |
Protocadherin-psi1; Uncharacterized protein |
| chr11_+_113779704 | 0.07 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr14_-_25103388 | 0.06 |
ENST00000526004.1 ENST00000415355.3 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chrX_+_95939711 | 0.06 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr7_+_74072011 | 0.04 |
ENST00000324896.4 ENST00000353920.4 ENST00000346152.4 ENST00000416070.1 |
GTF2I |
general transcription factor IIi |
| chr2_+_173686303 | 0.04 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr13_-_73356009 | 0.04 |
ENST00000377780.4 ENST00000377767.4 |
DIS3 |
DIS3 mitotic control homolog (S. cerevisiae) |
| chrX_-_54209640 | 0.04 |
ENST00000375180.2 ENST00000328235.4 ENST00000477084.1 |
FAM120C |
family with sequence similarity 120C |
| chr7_-_122840015 | 0.04 |
ENST00000194130.2 |
SLC13A1 |
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr18_+_55018044 | 0.02 |
ENST00000324000.3 |
ST8SIA3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr14_-_25103472 | 0.02 |
ENST00000216341.4 ENST00000382542.1 ENST00000382540.1 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr6_-_136788001 | 0.02 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr13_+_52436111 | 0.02 |
ENST00000242819.4 |
CCDC70 |
coiled-coil domain containing 70 |
| chr1_-_153521714 | 0.02 |
ENST00000368713.3 |
S100A3 |
S100 calcium binding protein A3 |
| chr17_-_76183111 | 0.02 |
ENST00000405273.1 ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1 |
thymidine kinase 1, soluble |
| chr9_-_113018835 | 0.01 |
ENST00000374517.5 |
TXN |
thioredoxin |
| chr19_-_7812446 | 0.01 |
ENST00000394173.4 ENST00000301357.8 |
CD209 |
CD209 molecule |
| chr16_-_66952779 | 0.01 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr13_-_33112899 | 0.01 |
ENST00000267068.3 ENST00000357505.6 ENST00000399396.3 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr17_+_77021702 | 0.00 |
ENST00000392445.2 ENST00000354124.3 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr17_+_66539369 | 0.00 |
ENST00000600820.1 |
AC079210.1 |
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.3 | 3.0 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.8 | GO:0086077 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.2 | 1.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.2 | GO:0015217 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 3.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 7.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 5.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.7 | 5.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.6 | 4.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 3.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.4 | 1.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.3 | 1.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.3 | 0.8 | GO:1903410 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.3 | 0.8 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.3 | 3.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.7 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 0.8 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.2 | 0.7 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.9 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 3.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 1.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.2 | GO:1904434 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.6 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 3.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 3.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 3.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 5.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0016528 | sarcoplasm(GO:0016528) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 4.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 8.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |