Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
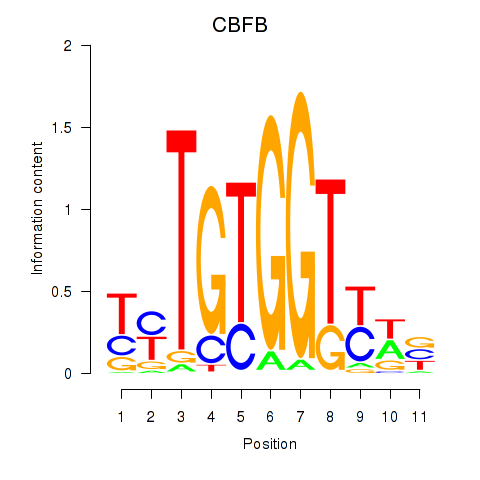
Results for CBFB
Z-value: 0.68
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | CBFB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67062996_67063019 | -0.73 | 1.4e-03 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_157154578 | 2.49 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr11_+_114310237 | 1.83 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr3_-_99569821 | 1.83 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr11_+_114310102 | 1.78 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr5_+_35856951 | 1.75 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr11_+_114310164 | 1.74 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr14_+_85996471 | 1.60 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_196621002 | 1.09 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr1_+_196621156 | 0.87 |
ENST00000359637.2 |
CFH |
complement factor H |
| chr14_+_85996507 | 0.87 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_32582293 | 0.85 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr14_-_61116168 | 0.65 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr1_-_240775447 | 0.63 |
ENST00000318160.4 |
GREM2 |
gremlin 2, DAN family BMP antagonist |
| chr4_-_156297949 | 0.60 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr20_+_43343886 | 0.58 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr18_-_53089723 | 0.51 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr3_-_189840223 | 0.47 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr21_-_36421535 | 0.47 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr21_-_36421626 | 0.47 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr11_+_18343800 | 0.47 |
ENST00000453096.2 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr5_-_137878887 | 0.43 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr11_-_58343319 | 0.43 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr4_+_74347400 | 0.42 |
ENST00000226355.3 |
AFM |
afamin |
| chr10_-_29811456 | 0.39 |
ENST00000535393.1 |
SVIL |
supervillin |
| chr12_-_68553512 | 0.34 |
ENST00000229135.3 |
IFNG |
interferon, gamma |
| chr1_+_209859510 | 0.31 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr2_-_87248975 | 0.29 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr20_+_54987168 | 0.29 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr15_+_49715293 | 0.26 |
ENST00000267843.4 ENST00000560270.1 |
FGF7 |
fibroblast growth factor 7 |
| chr7_+_120628731 | 0.24 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr20_+_54987305 | 0.23 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr21_-_36421401 | 0.22 |
ENST00000486278.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr12_-_14849470 | 0.21 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr9_+_15422702 | 0.18 |
ENST00000380821.3 ENST00000421710.1 |
SNAPC3 |
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr11_-_66103867 | 0.18 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr1_-_184723942 | 0.16 |
ENST00000318130.8 |
EDEM3 |
ER degradation enhancer, mannosidase alpha-like 3 |
| chr16_+_53088885 | 0.15 |
ENST00000566029.1 ENST00000447540.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr5_+_131409476 | 0.13 |
ENST00000296871.2 |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
| chr9_-_35080013 | 0.13 |
ENST00000378643.3 |
FANCG |
Fanconi anemia, complementation group G |
| chrX_-_153718988 | 0.12 |
ENST00000263512.4 ENST00000393587.4 ENST00000453912.1 |
SLC10A3 |
solute carrier family 10, member 3 |
| chrX_+_68048803 | 0.10 |
ENST00000204961.4 |
EFNB1 |
ephrin-B1 |
| chr5_+_131396222 | 0.10 |
ENST00000296870.2 |
IL3 |
interleukin 3 (colony-stimulating factor, multiple) |
| chr11_-_18343669 | 0.09 |
ENST00000396253.3 ENST00000349215.3 ENST00000438420.2 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
| chr8_-_60031762 | 0.08 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr11_-_64885111 | 0.08 |
ENST00000528598.1 ENST00000310597.4 |
ZNHIT2 |
zinc finger, HIT-type containing 2 |
| chr2_+_207024306 | 0.07 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_179112189 | 0.06 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr14_+_22919081 | 0.05 |
ENST00000390473.1 |
TRDJ1 |
T cell receptor delta joining 1 |
| chr6_+_27925019 | 0.04 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr15_+_64386261 | 0.04 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr14_-_24977457 | 0.03 |
ENST00000250378.3 ENST00000206446.4 |
CMA1 |
chymase 1, mast cell |
| chr6_+_29068386 | 0.03 |
ENST00000377171.3 |
OR2J1 |
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr9_-_123342415 | 0.02 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr13_-_28674693 | 0.00 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr1_+_89246647 | 0.00 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 5.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 2.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.4 | GO:0072562 | blood microparticle(GO:0072562) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.4 | 2.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 0.3 | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.3 | 0.8 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.2 | 0.6 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.6 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 1.8 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 5.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:0045918 | neutrophil differentiation(GO:0030223) negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 2.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 2.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 5.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 2.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |