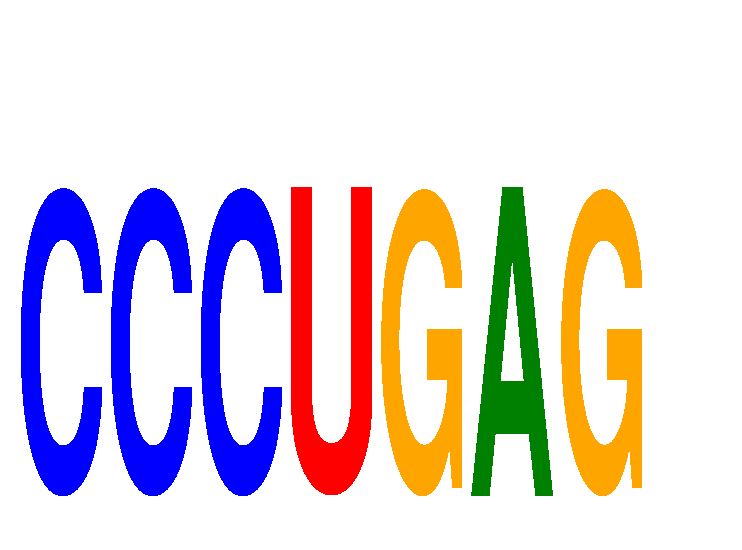Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for CCCUGAG
Z-value: 1.64
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-125a-5p
|
MIMAT0000443 |
|
hsa-miR-125b-5p
|
MIMAT0000423 |
|
hsa-miR-4319
|
MIMAT0016870 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_61521495 | 3.43 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr6_+_138188551 | 3.31 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr3_+_186648274 | 2.93 |
ENST00000169298.3 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_+_926000 | 2.79 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr2_+_7057523 | 2.75 |
ENST00000320892.6 |
RNF144A |
ring finger protein 144A |
| chr12_-_125348448 | 2.58 |
ENST00000339570.5 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr8_-_8751068 | 2.54 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr7_-_22233442 | 2.53 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_-_57301748 | 2.37 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr12_+_56473628 | 2.34 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr6_-_16761678 | 2.16 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr15_+_74833518 | 2.11 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_-_13461807 | 2.09 |
ENST00000254508.5 |
NUP210 |
nucleoporin 210kDa |
| chr17_+_55162453 | 1.94 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr22_+_21771656 | 1.91 |
ENST00000407464.2 |
HIC2 |
hypermethylated in cancer 2 |
| chr12_-_63328817 | 1.90 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr5_-_175964366 | 1.88 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr2_+_75061108 | 1.66 |
ENST00000290573.2 |
HK2 |
hexokinase 2 |
| chr2_-_201828356 | 1.58 |
ENST00000234296.2 |
ORC2 |
origin recognition complex, subunit 2 |
| chr1_+_26856236 | 1.57 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr6_+_106546808 | 1.54 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr14_-_90085458 | 1.54 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr6_+_391739 | 1.45 |
ENST00000380956.4 |
IRF4 |
interferon regulatory factor 4 |
| chr3_-_48672859 | 1.44 |
ENST00000395550.2 ENST00000455886.2 ENST00000431739.1 ENST00000426599.1 ENST00000383733.3 ENST00000420764.2 ENST00000337000.8 |
SLC26A6 |
solute carrier family 26 (anion exchanger), member 6 |
| chr7_-_139876812 | 1.43 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr11_-_102323489 | 1.40 |
ENST00000361236.3 |
TMEM123 |
transmembrane protein 123 |
| chr17_-_66287257 | 1.37 |
ENST00000327268.4 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr17_-_40540377 | 1.36 |
ENST00000404395.3 ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr3_-_72496035 | 1.34 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr18_-_60987220 | 1.33 |
ENST00000398117.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr17_+_80477571 | 1.29 |
ENST00000335255.5 |
FOXK2 |
forkhead box K2 |
| chr2_-_97535708 | 1.27 |
ENST00000305476.5 |
SEMA4C |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr11_+_75526212 | 1.27 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr12_+_11802753 | 1.27 |
ENST00000396373.4 |
ETV6 |
ets variant 6 |
| chr8_-_28243934 | 1.26 |
ENST00000521185.1 ENST00000520290.1 ENST00000344423.5 |
ZNF395 |
zinc finger protein 395 |
| chr18_-_19284724 | 1.22 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr1_+_214161272 | 1.20 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr17_-_76713100 | 1.17 |
ENST00000585509.1 |
CYTH1 |
cytohesin 1 |
| chr17_-_5372271 | 1.15 |
ENST00000225296.3 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr14_-_91976488 | 1.14 |
ENST00000554684.1 ENST00000337238.4 ENST00000428424.2 ENST00000554511.1 |
SMEK1 |
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr16_+_66400533 | 1.13 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr6_-_31830655 | 1.11 |
ENST00000375631.4 |
NEU1 |
sialidase 1 (lysosomal sialidase) |
| chr20_+_55966444 | 1.11 |
ENST00000356208.5 ENST00000440234.2 |
RBM38 |
RNA binding motif protein 38 |
| chr15_+_81489213 | 1.10 |
ENST00000559383.1 ENST00000394660.2 |
IL16 |
interleukin 16 |
| chr11_-_65381643 | 1.09 |
ENST00000309100.3 ENST00000529839.1 ENST00000526293.1 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
| chr6_-_90062543 | 1.09 |
ENST00000435041.2 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
| chr1_-_108507631 | 1.07 |
ENST00000527011.1 ENST00000370056.4 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr20_+_5107420 | 1.06 |
ENST00000460006.1 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr18_-_57364588 | 1.04 |
ENST00000439986.4 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
| chr11_+_63706444 | 1.03 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr22_-_39548627 | 1.02 |
ENST00000216133.5 |
CBX7 |
chromobox homolog 7 |
| chr3_+_58223228 | 1.02 |
ENST00000478253.1 ENST00000295962.4 |
ABHD6 |
abhydrolase domain containing 6 |
| chr14_+_74111578 | 1.01 |
ENST00000554113.1 ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1 |
dynein, axonemal, light chain 1 |
| chr10_+_97515409 | 0.98 |
ENST00000371207.3 ENST00000543964.1 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr17_-_40761375 | 0.97 |
ENST00000543197.1 ENST00000309428.5 |
FAM134C |
family with sequence similarity 134, member C |
| chr3_+_183353356 | 0.96 |
ENST00000242810.6 ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24 |
kelch-like family member 24 |
| chr9_-_100935043 | 0.96 |
ENST00000343933.5 |
CORO2A |
coronin, actin binding protein, 2A |
| chr18_+_55102917 | 0.95 |
ENST00000491143.2 |
ONECUT2 |
one cut homeobox 2 |
| chr3_-_53381539 | 0.95 |
ENST00000606822.1 ENST00000294241.6 ENST00000607628.1 |
DCP1A |
decapping mRNA 1A |
| chr17_-_61777459 | 0.94 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr20_-_32262165 | 0.93 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chrX_+_24483338 | 0.93 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr6_+_7107999 | 0.93 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr6_-_111136513 | 0.92 |
ENST00000368911.3 |
CDK19 |
cyclin-dependent kinase 19 |
| chr10_-_118764862 | 0.92 |
ENST00000260777.10 |
KIAA1598 |
KIAA1598 |
| chr19_-_2050852 | 0.90 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr2_+_61293021 | 0.90 |
ENST00000402291.1 |
KIAA1841 |
KIAA1841 |
| chr1_+_89990431 | 0.86 |
ENST00000330947.2 ENST00000358200.4 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
| chr4_+_38665810 | 0.84 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr7_+_49813255 | 0.84 |
ENST00000340652.4 |
VWC2 |
von Willebrand factor C domain containing 2 |
| chr9_-_130829588 | 0.83 |
ENST00000373078.4 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
| chr16_-_23521710 | 0.83 |
ENST00000562117.1 ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr3_+_51575596 | 0.82 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr16_-_19729502 | 0.82 |
ENST00000219837.7 |
KNOP1 |
lysine-rich nucleolar protein 1 |
| chr14_+_70078303 | 0.82 |
ENST00000342745.4 |
KIAA0247 |
KIAA0247 |
| chr22_-_50699701 | 0.81 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr3_+_38495333 | 0.79 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr2_+_177053307 | 0.78 |
ENST00000331462.4 |
HOXD1 |
homeobox D1 |
| chr17_+_53828381 | 0.77 |
ENST00000576183.1 |
PCTP |
phosphatidylcholine transfer protein |
| chr18_-_29264669 | 0.77 |
ENST00000306851.5 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr12_+_5019061 | 0.77 |
ENST00000382545.3 |
KCNA1 |
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr18_+_32558208 | 0.75 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr20_-_4982132 | 0.75 |
ENST00000338244.1 ENST00000424750.2 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr2_-_201936302 | 0.75 |
ENST00000453765.1 ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B |
family with sequence similarity 126, member B |
| chr6_-_11232891 | 0.74 |
ENST00000379433.5 ENST00000379446.5 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_-_207224307 | 0.74 |
ENST00000315927.4 |
YOD1 |
YOD1 deubiquitinase |
| chr11_+_111945011 | 0.72 |
ENST00000532163.1 ENST00000280352.9 ENST00000530104.1 ENST00000526879.1 ENST00000393047.3 ENST00000525785.1 |
C11orf57 |
chromosome 11 open reading frame 57 |
| chr4_+_37892682 | 0.72 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr1_-_36022979 | 0.72 |
ENST00000469892.1 ENST00000325722.3 |
KIAA0319L |
KIAA0319-like |
| chr10_-_15762124 | 0.72 |
ENST00000378076.3 |
ITGA8 |
integrin, alpha 8 |
| chr16_+_68298405 | 0.71 |
ENST00000219343.6 ENST00000566834.1 ENST00000566454.1 |
SLC7A6 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr3_+_47021168 | 0.71 |
ENST00000450053.3 ENST00000292309.5 ENST00000383740.2 |
NBEAL2 |
neurobeachin-like 2 |
| chrX_+_146993449 | 0.68 |
ENST00000218200.8 ENST00000370471.3 ENST00000370477.1 |
FMR1 |
fragile X mental retardation 1 |
| chr12_-_57030115 | 0.68 |
ENST00000379441.3 ENST00000179765.5 ENST00000551812.1 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
| chr11_-_119252359 | 0.67 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr10_-_94003003 | 0.67 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr11_-_46940074 | 0.66 |
ENST00000378623.1 ENST00000534404.1 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
| chr16_+_71929397 | 0.66 |
ENST00000537613.1 ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
| chr9_-_35650900 | 0.65 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr19_-_4066890 | 0.65 |
ENST00000322357.4 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
| chr18_+_18943554 | 0.63 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr15_-_93199069 | 0.62 |
ENST00000327355.5 |
FAM174B |
family with sequence similarity 174, member B |
| chr1_+_165796753 | 0.62 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr11_+_58939965 | 0.61 |
ENST00000227451.3 |
DTX4 |
deltex homolog 4 (Drosophila) |
| chr5_+_96271141 | 0.61 |
ENST00000231368.5 |
LNPEP |
leucyl/cystinyl aminopeptidase |
| chr6_+_132129151 | 0.61 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr6_+_138483058 | 0.60 |
ENST00000251691.4 |
KIAA1244 |
KIAA1244 |
| chr20_-_52790512 | 0.60 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr14_+_60715928 | 0.59 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr3_+_171758344 | 0.59 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr15_+_31619013 | 0.58 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr11_+_86748863 | 0.57 |
ENST00000340353.7 |
TMEM135 |
transmembrane protein 135 |
| chr19_-_49944806 | 0.55 |
ENST00000221485.3 |
SLC17A7 |
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr8_+_76452097 | 0.54 |
ENST00000396423.2 |
HNF4G |
hepatocyte nuclear factor 4, gamma |
| chr20_-_30795511 | 0.54 |
ENST00000246229.4 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
| chr2_+_208394616 | 0.54 |
ENST00000432329.2 ENST00000353267.3 ENST00000445803.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr6_-_24911195 | 0.53 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr2_-_200322723 | 0.52 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr19_-_19754404 | 0.51 |
ENST00000587205.1 ENST00000445806.2 ENST00000203556.4 |
GMIP |
GEM interacting protein |
| chr22_-_31364187 | 0.50 |
ENST00000215862.4 ENST00000397641.3 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr19_+_11485333 | 0.49 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr4_+_57774042 | 0.49 |
ENST00000309042.7 |
REST |
RE1-silencing transcription factor |
| chr2_-_206950781 | 0.49 |
ENST00000403263.1 |
INO80D |
INO80 complex subunit D |
| chr12_-_56727487 | 0.48 |
ENST00000548043.1 ENST00000425394.2 |
PAN2 |
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_+_141205852 | 0.48 |
ENST00000286364.3 ENST00000452898.1 |
RASA2 |
RAS p21 protein activator 2 |
| chr17_+_41177220 | 0.48 |
ENST00000587250.2 ENST00000544533.1 |
RND2 |
Rho family GTPase 2 |
| chr6_-_33547975 | 0.47 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr8_-_145550571 | 0.47 |
ENST00000332324.4 |
DGAT1 |
diacylglycerol O-acyltransferase 1 |
| chr1_+_12227035 | 0.47 |
ENST00000376259.3 ENST00000536782.1 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
| chr1_+_203274639 | 0.47 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr22_+_42229100 | 0.47 |
ENST00000361204.4 |
SREBF2 |
sterol regulatory element binding transcription factor 2 |
| chr7_+_142985308 | 0.47 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr1_+_109792641 | 0.46 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr2_-_103353277 | 0.46 |
ENST00000258436.5 |
MFSD9 |
major facilitator superfamily domain containing 9 |
| chr21_-_40685477 | 0.46 |
ENST00000342449.3 |
BRWD1 |
bromodomain and WD repeat domain containing 1 |
| chr10_+_97803151 | 0.44 |
ENST00000403870.3 ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ |
cyclin J |
| chr20_-_524455 | 0.44 |
ENST00000349736.5 ENST00000217244.3 |
CSNK2A1 |
casein kinase 2, alpha 1 polypeptide |
| chr5_-_100238956 | 0.43 |
ENST00000231461.5 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr2_+_204192942 | 0.43 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr16_-_18937726 | 0.43 |
ENST00000389467.3 ENST00000446231.2 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr17_-_38020392 | 0.42 |
ENST00000346872.3 ENST00000439167.2 ENST00000377945.3 ENST00000394189.2 ENST00000377944.3 ENST00000377958.2 ENST00000535189.1 ENST00000377952.2 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
| chr16_+_8768422 | 0.42 |
ENST00000268251.8 ENST00000564714.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr4_-_41750922 | 0.41 |
ENST00000226382.2 |
PHOX2B |
paired-like homeobox 2b |
| chr12_-_42538657 | 0.41 |
ENST00000398675.3 |
GXYLT1 |
glucoside xylosyltransferase 1 |
| chr4_+_75858290 | 0.40 |
ENST00000513238.1 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr6_+_24667257 | 0.40 |
ENST00000537591.1 ENST00000230048.4 |
ACOT13 |
acyl-CoA thioesterase 13 |
| chr11_-_78052923 | 0.40 |
ENST00000340149.2 |
GAB2 |
GRB2-associated binding protein 2 |
| chr19_-_47975417 | 0.39 |
ENST00000236877.6 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr22_+_29601840 | 0.39 |
ENST00000334018.6 ENST00000429226.1 ENST00000404755.3 ENST00000404820.3 ENST00000430127.1 |
EMID1 |
EMI domain containing 1 |
| chr5_+_56111361 | 0.39 |
ENST00000399503.3 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr10_+_73975742 | 0.38 |
ENST00000299381.4 |
ANAPC16 |
anaphase promoting complex subunit 16 |
| chr11_-_128392085 | 0.38 |
ENST00000526145.2 ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr1_-_151319710 | 0.38 |
ENST00000290524.4 ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr20_+_62795827 | 0.37 |
ENST00000328439.1 ENST00000536311.1 |
MYT1 |
myelin transcription factor 1 |
| chr10_-_103454876 | 0.37 |
ENST00000331272.7 |
FBXW4 |
F-box and WD repeat domain containing 4 |
| chr3_+_5229356 | 0.36 |
ENST00000256497.4 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
| chr1_+_112162381 | 0.36 |
ENST00000433097.1 ENST00000369709.3 ENST00000436150.2 |
RAP1A |
RAP1A, member of RAS oncogene family |
| chr3_-_133614597 | 0.36 |
ENST00000285208.4 ENST00000460865.3 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr2_+_28615669 | 0.36 |
ENST00000379619.1 ENST00000264716.4 |
FOSL2 |
FOS-like antigen 2 |
| chr3_-_197282821 | 0.36 |
ENST00000445160.2 ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_+_232651124 | 0.35 |
ENST00000350033.3 ENST00000412591.1 ENST00000410017.1 ENST00000373608.3 |
COPS7B |
COP9 signalosome subunit 7B |
| chr2_-_26101374 | 0.34 |
ENST00000435504.4 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
| chr22_-_44258360 | 0.34 |
ENST00000330884.4 ENST00000249130.5 |
SULT4A1 |
sulfotransferase family 4A, member 1 |
| chr1_+_15573757 | 0.34 |
ENST00000358897.4 ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr1_+_50574585 | 0.33 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr2_-_85555385 | 0.33 |
ENST00000377386.3 |
TGOLN2 |
trans-golgi network protein 2 |
| chr3_+_183873098 | 0.33 |
ENST00000313143.3 |
DVL3 |
dishevelled segment polarity protein 3 |
| chr16_-_71610985 | 0.33 |
ENST00000355962.4 |
TAT |
tyrosine aminotransferase |
| chr19_+_19303008 | 0.32 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr16_+_67063036 | 0.32 |
ENST00000290858.6 ENST00000564034.1 |
CBFB |
core-binding factor, beta subunit |
| chr13_-_27745936 | 0.32 |
ENST00000282344.6 |
USP12 |
ubiquitin specific peptidase 12 |
| chr10_+_82168240 | 0.32 |
ENST00000372187.5 ENST00000372185.1 |
FAM213A |
family with sequence similarity 213, member A |
| chr2_+_131113580 | 0.32 |
ENST00000175756.5 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
| chr13_-_30169807 | 0.32 |
ENST00000380752.5 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_-_120806960 | 0.32 |
ENST00000257552.2 |
MSI1 |
musashi RNA-binding protein 1 |
| chr1_-_217262969 | 0.31 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr3_+_9773409 | 0.31 |
ENST00000433861.2 ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1 |
bromodomain and PHD finger containing, 1 |
| chr3_+_15468862 | 0.31 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr1_-_109940550 | 0.30 |
ENST00000256637.6 |
SORT1 |
sortilin 1 |
| chr9_-_3525968 | 0.30 |
ENST00000382004.3 ENST00000302303.1 ENST00000449190.1 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
| chr19_-_41256207 | 0.30 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr19_+_4969116 | 0.29 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr11_+_118992269 | 0.29 |
ENST00000350777.2 ENST00000529988.1 ENST00000527410.1 |
HINFP |
histone H4 transcription factor |
| chr20_+_39765581 | 0.28 |
ENST00000244007.3 |
PLCG1 |
phospholipase C, gamma 1 |
| chr5_+_110559784 | 0.28 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr12_+_49761224 | 0.28 |
ENST00000553127.1 ENST00000321898.6 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
| chr3_+_141106643 | 0.28 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chrX_-_19988382 | 0.28 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr6_+_35995488 | 0.28 |
ENST00000229795.3 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr15_+_85923856 | 0.27 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chrX_+_153639856 | 0.27 |
ENST00000426834.1 ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ |
tafazzin |
| chr6_-_32811771 | 0.27 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr9_-_116102530 | 0.27 |
ENST00000374195.3 ENST00000341761.4 |
WDR31 |
WD repeat domain 31 |
| chr2_+_176987088 | 0.26 |
ENST00000249499.6 |
HOXD9 |
homeobox D9 |
| chr20_+_306221 | 0.26 |
ENST00000342665.2 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chrX_-_48755030 | 0.26 |
ENST00000490755.2 ENST00000465150.2 ENST00000495490.2 |
TIMM17B |
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr10_+_14920843 | 0.26 |
ENST00000433779.1 ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr12_-_51717922 | 0.26 |
ENST00000452142.2 |
BIN2 |
bridging integrator 2 |
| chr12_+_109554386 | 0.25 |
ENST00000338432.7 |
ACACB |
acetyl-CoA carboxylase beta |
| chr15_+_90544532 | 0.25 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 3.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 2.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.6 | 2.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.6 | 2.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.5 | 3.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 4.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 1.4 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.4 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.2 | 0.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.7 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.0 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.2 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 2.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.1 | 1.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.6 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.8 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 1.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 2.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 2.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.7 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 5.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.4 | 2.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.2 | 0.7 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.2 | 0.7 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.2 | 0.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 1.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.6 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 1.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 4.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.8 | GO:0034702 | ion channel complex(GO:0034702) transmembrane transporter complex(GO:1902495) transporter complex(GO:1990351) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.2 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.2 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 4.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 5.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.9 | 2.6 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.5 | 1.4 | GO:0045082 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.5 | 1.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.5 | 2.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 1.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.4 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.3 | 1.3 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.3 | 1.5 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.2 | 0.7 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.7 | GO:1904925 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 0.9 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 3.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 2.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 0.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.2 | 0.7 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 | 1.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 1.0 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.2 | 1.0 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.6 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.2 | 1.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.2 | 0.9 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 0.6 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.2 | 1.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.5 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 0.5 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.6 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 0.9 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 1.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.4 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.2 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.8 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.8 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.4 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.6 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 0.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 2.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.5 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 1.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.9 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.2 | GO:0098904 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.5 | GO:0035234 | luteolysis(GO:0001554) ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.5 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.8 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.6 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.0 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.1 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 1.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 2.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.2 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 0.8 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.2 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.3 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 1.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.5 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 2.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 1.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 1.0 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0046676 | negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.7 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.8 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 3.3 | GO:1902400 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.3 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0030948 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 1.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 1.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.7 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.4 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.2 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 2.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 1.0 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.4 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.7 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0051582 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.8 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.7 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.0 | GO:0044256 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0050779 | RNA destabilization(GO:0050779) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |