Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
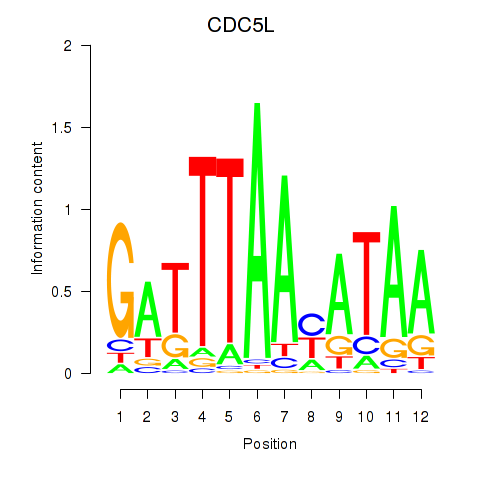
Results for CDC5L
Z-value: 1.08
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | CDC5L |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | 0.29 | 2.8e-01 | Click! |
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_127413481 | 3.22 |
ENST00000259254.4 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr6_-_49604545 | 2.02 |
ENST00000371175.4 ENST00000229810.7 |
RHAG |
Rh-associated glycoprotein |
| chr1_+_198608146 | 1.48 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr15_-_55562582 | 1.43 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55562479 | 1.42 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_-_53601055 | 1.31 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr6_-_32557610 | 1.26 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr15_-_55611306 | 1.14 |
ENST00000563262.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_+_118690466 | 1.12 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_13182751 | 1.11 |
ENST00000415087.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr12_-_53601000 | 1.04 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr11_+_59824060 | 1.03 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr7_+_116654935 | 1.02 |
ENST00000432298.1 ENST00000422922.1 |
ST7 |
suppression of tumorigenicity 7 |
| chrX_+_52780318 | 0.99 |
ENST00000375515.3 ENST00000276049.6 |
SSX2B |
synovial sarcoma, X breakpoint 2B |
| chrX_+_65382433 | 0.99 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr8_+_99956662 | 0.99 |
ENST00000523368.1 ENST00000297565.4 ENST00000435298.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr6_-_32498046 | 0.98 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr11_+_59824127 | 0.97 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr6_+_32812568 | 0.95 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr16_+_33204156 | 0.89 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr5_-_133510456 | 0.89 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr13_-_38172863 | 0.85 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chrX_+_30261847 | 0.83 |
ENST00000378981.3 ENST00000397550.1 |
MAGEB1 |
melanoma antigen family B, 1 |
| chr22_+_19467261 | 0.82 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chr3_+_20081515 | 0.81 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr10_-_69597810 | 0.81 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr3_-_119278446 | 0.79 |
ENST00000264246.3 |
CD80 |
CD80 molecule |
| chr3_-_119278376 | 0.79 |
ENST00000478182.1 |
CD80 |
CD80 molecule |
| chr6_+_32709119 | 0.78 |
ENST00000374940.3 |
HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
| chr11_-_118213331 | 0.77 |
ENST00000392884.2 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr8_-_86253888 | 0.77 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr14_-_21516590 | 0.77 |
ENST00000555026.1 |
NDRG2 |
NDRG family member 2 |
| chr1_-_150738261 | 0.76 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chrX_-_48056199 | 0.75 |
ENST00000311798.1 ENST00000347757.1 |
SSX5 |
synovial sarcoma, X breakpoint 5 |
| chr22_+_19466980 | 0.74 |
ENST00000407835.1 ENST00000438587.1 |
CDC45 |
cell division cycle 45 |
| chr22_-_39268308 | 0.73 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr6_+_35996859 | 0.72 |
ENST00000472333.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr6_+_57182400 | 0.71 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr3_+_178276488 | 0.70 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr10_-_75226166 | 0.69 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr1_+_25598989 | 0.69 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chrX_-_48216101 | 0.68 |
ENST00000298396.2 ENST00000376893.3 |
SSX3 |
synovial sarcoma, X breakpoint 3 |
| chr11_+_125496619 | 0.68 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chrX_-_23926004 | 0.68 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr13_-_47012325 | 0.67 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr10_+_91152303 | 0.67 |
ENST00000371804.3 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 |
| chr11_-_118213455 | 0.67 |
ENST00000300692.4 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr1_+_241695670 | 0.66 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr6_-_33041378 | 0.65 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr10_-_98031310 | 0.65 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr9_+_135854091 | 0.65 |
ENST00000450530.1 ENST00000534944.1 |
GFI1B |
growth factor independent 1B transcription repressor |
| chr19_-_53662257 | 0.64 |
ENST00000599096.1 ENST00000334197.7 ENST00000597183.1 ENST00000601804.1 ENST00000601469.2 ENST00000452676.2 |
ZNF347 |
zinc finger protein 347 |
| chr14_-_106725723 | 0.64 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr2_+_232573208 | 0.62 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr12_+_58176525 | 0.61 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr6_-_39197226 | 0.61 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr5_+_141346385 | 0.61 |
ENST00000513019.1 ENST00000356143.1 |
RNF14 |
ring finger protein 14 |
| chr1_+_95616933 | 0.60 |
ENST00000604203.1 |
RP11-57H12.6 |
TMEM56-RWDD3 readthrough |
| chr18_-_31628558 | 0.59 |
ENST00000535384.1 |
NOL4 |
nucleolar protein 4 |
| chr10_-_98031265 | 0.59 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr11_-_62609281 | 0.56 |
ENST00000525239.1 ENST00000538098.2 |
WDR74 |
WD repeat domain 74 |
| chr1_+_196621156 | 0.56 |
ENST00000359637.2 |
CFH |
complement factor H |
| chr14_-_106733624 | 0.55 |
ENST00000390610.2 |
IGHV1-24 |
immunoglobulin heavy variable 1-24 |
| chr1_-_25747283 | 0.54 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr12_-_21927736 | 0.54 |
ENST00000240662.2 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr1_+_43291220 | 0.54 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr11_-_118213360 | 0.53 |
ENST00000529594.1 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr1_-_247094628 | 0.53 |
ENST00000366508.1 ENST00000326225.3 ENST00000391829.2 |
AHCTF1 |
AT hook containing transcription factor 1 |
| chr2_+_232573222 | 0.53 |
ENST00000341369.7 ENST00000409683.1 |
PTMA |
prothymosin, alpha |
| chr6_+_26156551 | 0.53 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr19_+_30433110 | 0.51 |
ENST00000542441.2 ENST00000392271.1 |
URI1 |
URI1, prefoldin-like chaperone |
| chr1_+_70820451 | 0.50 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr12_-_9913489 | 0.50 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr7_+_77469439 | 0.50 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr21_-_33975547 | 0.49 |
ENST00000431599.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr3_+_69928256 | 0.48 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_100316041 | 0.48 |
ENST00000370165.3 ENST00000370163.3 ENST00000294724.4 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr18_+_61554932 | 0.48 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr4_+_80584903 | 0.47 |
ENST00000506460.1 |
RP11-452C8.1 |
RP11-452C8.1 |
| chr10_-_69597915 | 0.47 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr9_+_125132803 | 0.47 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_-_20193370 | 0.46 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr10_-_62493223 | 0.45 |
ENST00000373827.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr14_-_106668095 | 0.45 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr8_+_11666649 | 0.45 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_-_176914238 | 0.44 |
ENST00000430069.1 ENST00000428970.1 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
| chr22_-_29107919 | 0.43 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr1_+_25599018 | 0.41 |
ENST00000417538.2 ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD |
Rh blood group, D antigen |
| chr12_-_91546926 | 0.41 |
ENST00000550758.1 |
DCN |
decorin |
| chr6_-_26250835 | 0.41 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr11_+_2415061 | 0.41 |
ENST00000481687.1 |
CD81 |
CD81 molecule |
| chr12_-_91573249 | 0.39 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr8_+_27950619 | 0.39 |
ENST00000542181.1 ENST00000524103.1 ENST00000537665.1 ENST00000380353.4 ENST00000520288.1 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
| chr4_-_38806404 | 0.39 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr7_+_116593433 | 0.39 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr15_+_35270552 | 0.39 |
ENST00000391457.2 |
AC114546.1 |
HCG37415; PRO1914; Uncharacterized protein |
| chr9_-_110251836 | 0.39 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr7_+_129906660 | 0.38 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chr2_+_185463093 | 0.38 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr6_-_136847610 | 0.38 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr10_+_35484053 | 0.37 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr12_+_21525818 | 0.37 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr1_+_241695424 | 0.36 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_166095898 | 0.36 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_-_100356844 | 0.36 |
ENST00000437033.2 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_136847099 | 0.36 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr4_+_147096837 | 0.36 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_+_106631966 | 0.36 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr20_-_16554078 | 0.35 |
ENST00000354981.2 ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B |
kinesin family member 16B |
| chrX_+_77166172 | 0.35 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr1_+_196621002 | 0.35 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr12_-_54653313 | 0.35 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr8_-_101962777 | 0.35 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_+_150321068 | 0.34 |
ENST00000471696.1 ENST00000477889.1 ENST00000485923.1 |
SELT |
Selenoprotein T |
| chr17_-_2996290 | 0.33 |
ENST00000331459.1 |
OR1D2 |
olfactory receptor, family 1, subfamily D, member 2 |
| chr3_-_119396193 | 0.33 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr4_-_153274078 | 0.33 |
ENST00000263981.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr5_+_140213815 | 0.33 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr12_-_87232644 | 0.32 |
ENST00000549405.2 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_44001172 | 0.32 |
ENST00000260605.8 ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1 |
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr11_+_2405833 | 0.32 |
ENST00000527343.1 ENST00000464784.2 |
CD81 |
CD81 molecule |
| chr6_-_133055815 | 0.32 |
ENST00000509351.1 ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3 |
vanin 3 |
| chr19_-_17185848 | 0.32 |
ENST00000593360.1 |
HAUS8 |
HAUS augmin-like complex, subunit 8 |
| chr2_+_234601512 | 0.32 |
ENST00000305139.6 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr6_-_149969871 | 0.32 |
ENST00000335643.8 ENST00000444282.1 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr3_+_39424828 | 0.32 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr9_+_125133315 | 0.31 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_14026063 | 0.31 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr14_+_39703112 | 0.31 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr3_+_122044084 | 0.31 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr19_-_14889349 | 0.31 |
ENST00000315576.3 ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr16_+_1728257 | 0.30 |
ENST00000248098.3 ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr15_-_22448819 | 0.30 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr6_-_149969829 | 0.30 |
ENST00000367411.2 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr4_+_170541835 | 0.30 |
ENST00000504131.2 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr2_-_225811747 | 0.30 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr22_-_19466683 | 0.29 |
ENST00000399523.1 ENST00000421968.2 ENST00000447868.1 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr11_-_236326 | 0.29 |
ENST00000525237.1 ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3 |
sirtuin 3 |
| chr18_+_61442629 | 0.29 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr16_+_1728305 | 0.29 |
ENST00000569765.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr7_-_14026123 | 0.29 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr6_+_30297306 | 0.29 |
ENST00000420746.1 ENST00000513556.1 |
TRIM39 TRIM39-RPP21 |
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr19_-_4540486 | 0.28 |
ENST00000306390.6 |
LRG1 |
leucine-rich alpha-2-glycoprotein 1 |
| chr12_-_91573316 | 0.28 |
ENST00000393155.1 |
DCN |
decorin |
| chr18_-_57027194 | 0.28 |
ENST00000251047.5 |
LMAN1 |
lectin, mannose-binding, 1 |
| chr8_+_75736761 | 0.28 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr2_+_166326157 | 0.28 |
ENST00000421875.1 ENST00000314499.7 ENST00000409664.1 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr11_+_71249071 | 0.27 |
ENST00000398534.3 |
KRTAP5-8 |
keratin associated protein 5-8 |
| chr1_+_50571949 | 0.27 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr1_+_110254850 | 0.27 |
ENST00000369812.5 ENST00000256593.3 ENST00000369813.1 |
GSTM5 |
glutathione S-transferase mu 5 |
| chr5_+_33440802 | 0.27 |
ENST00000502553.1 ENST00000514259.1 ENST00000265112.3 |
TARS |
threonyl-tRNA synthetase |
| chr6_+_30312908 | 0.27 |
ENST00000433076.2 ENST00000442966.2 ENST00000428040.2 ENST00000436442.2 |
RPP21 |
ribonuclease P/MRP 21kDa subunit |
| chr1_-_9132311 | 0.27 |
ENST00000474145.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr22_-_39268192 | 0.27 |
ENST00000216083.6 |
CBX6 |
chromobox homolog 6 |
| chr7_+_80267973 | 0.26 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr19_-_1650666 | 0.26 |
ENST00000588136.1 |
TCF3 |
transcription factor 3 |
| chr12_-_21487829 | 0.26 |
ENST00000445053.1 ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2 |
solute carrier organic anion transporter family, member 1A2 |
| chr8_+_98900132 | 0.26 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr14_-_23877474 | 0.26 |
ENST00000405093.3 |
MYH6 |
myosin, heavy chain 6, cardiac muscle, alpha |
| chr16_-_4401258 | 0.26 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr13_+_111855414 | 0.25 |
ENST00000375737.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_+_65382381 | 0.25 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr3_-_118959733 | 0.25 |
ENST00000459778.1 ENST00000359213.3 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr1_+_174769006 | 0.25 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_-_231560790 | 0.25 |
ENST00000366641.3 |
EGLN1 |
egl-9 family hypoxia-inducible factor 1 |
| chr7_+_96634850 | 0.25 |
ENST00000518156.2 |
DLX6 |
distal-less homeobox 6 |
| chr4_-_76957214 | 0.24 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chrX_+_99839799 | 0.24 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr4_-_87028478 | 0.24 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr3_-_150320937 | 0.24 |
ENST00000479209.1 |
SERP1 |
stress-associated endoplasmic reticulum protein 1 |
| chr22_-_19466732 | 0.24 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr11_+_119076745 | 0.24 |
ENST00000264033.4 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr11_-_114466477 | 0.24 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr4_-_47465666 | 0.24 |
ENST00000381571.4 |
COMMD8 |
COMM domain containing 8 |
| chr4_-_38858428 | 0.24 |
ENST00000436693.2 ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6 TLR1 |
toll-like receptor 6 toll-like receptor 1 |
| chr1_+_41204506 | 0.24 |
ENST00000525290.1 ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC |
nuclear transcription factor Y, gamma |
| chr2_-_175462456 | 0.23 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr12_-_108955070 | 0.23 |
ENST00000228284.3 ENST00000546611.1 |
SART3 |
squamous cell carcinoma antigen recognized by T cells 3 |
| chr13_-_31038370 | 0.23 |
ENST00000399489.1 ENST00000339872.4 |
HMGB1 |
high mobility group box 1 |
| chr12_+_50794592 | 0.23 |
ENST00000293618.8 ENST00000429001.3 ENST00000548174.1 ENST00000548697.1 ENST00000548993.1 ENST00000398473.2 ENST00000522085.1 ENST00000518444.1 ENST00000551886.1 |
LARP4 |
La ribonucleoprotein domain family, member 4 |
| chr12_-_71182695 | 0.23 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr1_+_59775752 | 0.23 |
ENST00000371212.1 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr10_-_94257512 | 0.23 |
ENST00000371581.5 |
IDE |
insulin-degrading enzyme |
| chr2_-_233641265 | 0.23 |
ENST00000438786.1 ENST00000409779.1 ENST00000233826.3 |
KCNJ13 |
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr8_+_104831554 | 0.23 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr12_-_22063787 | 0.23 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_152956549 | 0.23 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr22_+_39077947 | 0.23 |
ENST00000216034.4 |
TOMM22 |
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr12_-_371994 | 0.22 |
ENST00000343164.4 ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13 |
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr11_-_83984231 | 0.22 |
ENST00000330014.6 ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr11_+_64794875 | 0.22 |
ENST00000377244.3 ENST00000534637.1 ENST00000524831.1 |
SNX15 |
sorting nexin 15 |
| chr20_+_43104508 | 0.22 |
ENST00000262605.4 ENST00000372904.3 |
TTPAL |
tocopherol (alpha) transfer protein-like |
| chr1_-_32169920 | 0.22 |
ENST00000373672.3 ENST00000373668.3 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr8_+_99956759 | 0.21 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chrX_+_100646190 | 0.21 |
ENST00000471855.1 |
RPL36A |
ribosomal protein L36a |
| chr18_-_31802282 | 0.21 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr9_-_93405352 | 0.21 |
ENST00000375765.3 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
| chr11_+_236959 | 0.21 |
ENST00000431206.2 ENST00000528906.1 |
PSMD13 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr3_+_190105909 | 0.21 |
ENST00000456423.1 |
CLDN16 |
claudin 16 |
| chr14_+_22520762 | 0.21 |
ENST00000390449.3 |
TRAV21 |
T cell receptor alpha variable 21 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.7 | 4.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.6 | 2.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.5 | 1.6 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 0.9 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.2 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 1.5 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.3 | 0.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 1.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.2 | 0.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.6 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.2 | 0.6 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 1.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 2.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.7 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.4 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.3 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.5 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.4 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.1 | 0.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.9 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 1.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0070318 | myoblast development(GO:0048627) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.4 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 2.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.3 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.2 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.3 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:2000317 | regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0009441 | fatty acid alpha-oxidation(GO:0001561) glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 1.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.6 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.7 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 2.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 | 0.6 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.3 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 1.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 3.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.4 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.2 | 0.6 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 4.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.5 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.1 | 1.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 2.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 2.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 1.7 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 3.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0052724 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) inositol hexakisphosphate kinase activity(GO:0000828) inositol heptakisphosphate kinase activity(GO:0000829) inositol hexakisphosphate 5-kinase activity(GO:0000832) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.0 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 3.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.5 | 1.6 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.3 | 4.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 0.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 2.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 3.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.9 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.3 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.1 | 2.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 1.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 2.5 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |