Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for CDX1
Z-value: 0.95
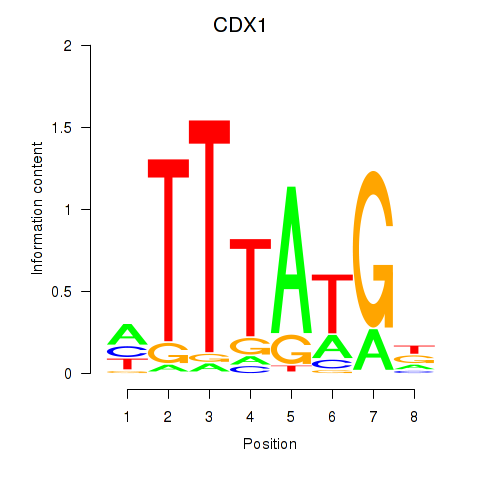
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | CDX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX1 | hg19_v2_chr5_+_149546334_149546364 | -0.17 | 5.4e-01 | Click! |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_149095652 | 4.99 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr2_-_88427568 | 4.17 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr12_-_9268707 | 3.91 |
ENST00000318602.7 |
A2M |
alpha-2-macroglobulin |
| chr4_-_110723194 | 2.40 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr22_+_21128167 | 2.21 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr4_-_110723335 | 2.12 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr3_+_190333097 | 2.10 |
ENST00000412080.1 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chr10_-_95360983 | 2.10 |
ENST00000371464.3 |
RBP4 |
retinol binding protein 4, plasma |
| chr3_-_123512688 | 2.05 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr11_-_2158507 | 2.02 |
ENST00000381392.1 ENST00000381395.1 ENST00000418738.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr10_-_52645379 | 2.02 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr4_+_74269956 | 1.98 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr10_-_52645416 | 1.97 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr16_+_56672571 | 1.91 |
ENST00000290705.8 |
MT1A |
metallothionein 1A |
| chr4_-_110723134 | 1.88 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr16_+_56685796 | 1.86 |
ENST00000334346.2 ENST00000562399.1 |
MT1B |
metallothionein 1B |
| chr10_-_69597915 | 1.64 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_-_90712520 | 1.62 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr9_+_124088860 | 1.58 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr20_+_56136136 | 1.57 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr4_-_16085314 | 1.46 |
ENST00000510224.1 |
PROM1 |
prominin 1 |
| chr4_-_16085340 | 1.39 |
ENST00000508167.1 |
PROM1 |
prominin 1 |
| chrX_-_131547625 | 1.36 |
ENST00000394311.2 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chrX_-_131547596 | 1.28 |
ENST00000538204.1 ENST00000370849.3 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr5_+_102201509 | 1.25 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr1_+_169079823 | 1.23 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr16_+_56691606 | 1.20 |
ENST00000334350.6 |
MT1F |
metallothionein 1F |
| chr10_-_69597810 | 1.19 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_+_81771806 | 1.11 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr18_+_3449695 | 1.09 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chrX_-_131623982 | 1.08 |
ENST00000370844.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr16_-_29910365 | 0.99 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr12_+_19358228 | 0.96 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr17_-_56492989 | 0.95 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chr5_-_111093759 | 0.94 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr5_+_102201430 | 0.92 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr4_+_71063641 | 0.92 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr1_+_89246647 | 0.89 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chr12_-_91573249 | 0.87 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr4_-_83765613 | 0.84 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr5_+_35856951 | 0.82 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr16_+_14927538 | 0.81 |
ENST00000287667.7 |
NOMO1 |
NODAL modulator 1 |
| chr12_-_91573132 | 0.80 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr17_-_41174424 | 0.76 |
ENST00000355653.3 |
VAT1 |
vesicle amine transport 1 |
| chr2_+_242289502 | 0.75 |
ENST00000451310.1 |
SEPT2 |
septin 2 |
| chr4_+_70146217 | 0.74 |
ENST00000335568.5 ENST00000511240.1 |
UGT2B28 |
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr5_-_16742330 | 0.73 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr15_+_80733570 | 0.73 |
ENST00000533983.1 ENST00000527771.1 ENST00000525103.1 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr9_-_21335356 | 0.69 |
ENST00000359039.4 |
KLHL9 |
kelch-like family member 9 |
| chr4_-_186732048 | 0.68 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr17_-_48943706 | 0.68 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr14_-_53331239 | 0.67 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr4_+_169418195 | 0.64 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr21_-_43735446 | 0.61 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr5_-_78281603 | 0.60 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr17_+_28705921 | 0.59 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr6_+_33168597 | 0.59 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr6_+_33168637 | 0.59 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr5_-_147211226 | 0.59 |
ENST00000296695.5 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr19_-_23578220 | 0.56 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr11_+_133938820 | 0.56 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr2_-_190927447 | 0.55 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr12_+_58087901 | 0.54 |
ENST00000315970.7 ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9 |
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr2_-_165424973 | 0.52 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr5_+_96079240 | 0.52 |
ENST00000515663.1 |
CAST |
calpastatin |
| chr6_+_158733692 | 0.51 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr21_-_43735628 | 0.49 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr6_+_143999072 | 0.49 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr2_+_161993412 | 0.47 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr12_+_51318513 | 0.47 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr2_+_32502952 | 0.47 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr11_+_114168085 | 0.46 |
ENST00000541754.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr4_+_169418255 | 0.46 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr5_+_140588269 | 0.45 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr10_-_63995871 | 0.45 |
ENST00000315289.2 |
RTKN2 |
rhotekin 2 |
| chr15_-_73925651 | 0.44 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr9_-_21335240 | 0.42 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr22_+_38004832 | 0.42 |
ENST00000405147.3 ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr5_+_140739537 | 0.41 |
ENST00000522605.1 |
PCDHGB2 |
protocadherin gamma subfamily B, 2 |
| chr2_+_161993465 | 0.40 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr3_+_135741576 | 0.40 |
ENST00000334546.2 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
| chr13_-_33780133 | 0.40 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr1_+_150337100 | 0.37 |
ENST00000401000.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr22_+_38004473 | 0.37 |
ENST00000414350.3 ENST00000343632.4 |
GGA1 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr3_-_49055991 | 0.37 |
ENST00000441576.2 ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3 |
DALR anticodon binding domain containing 3 |
| chr1_+_150337144 | 0.35 |
ENST00000539519.1 ENST00000369067.3 ENST00000369068.4 |
RPRD2 |
regulation of nuclear pre-mRNA domain containing 2 |
| chr6_+_36646435 | 0.34 |
ENST00000244741.5 ENST00000405375.1 ENST00000373711.2 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr2_-_208031943 | 0.34 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr21_+_44313375 | 0.33 |
ENST00000354250.2 ENST00000340344.4 |
NDUFV3 |
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr8_+_22423168 | 0.33 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr4_-_83769996 | 0.33 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr5_-_147211190 | 0.31 |
ENST00000510027.2 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr2_-_152589670 | 0.31 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr12_-_90049878 | 0.31 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_+_72080253 | 0.30 |
ENST00000549735.1 |
TMEM19 |
transmembrane protein 19 |
| chr2_+_46926048 | 0.29 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr6_+_26087509 | 0.27 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr6_-_43423308 | 0.27 |
ENST00000372485.1 ENST00000372488.3 |
DLK2 |
delta-like 2 homolog (Drosophila) |
| chr1_+_28764653 | 0.27 |
ENST00000373836.3 |
PHACTR4 |
phosphatase and actin regulator 4 |
| chr15_+_89164520 | 0.26 |
ENST00000332810.3 |
AEN |
apoptosis enhancing nuclease |
| chr8_-_42065187 | 0.26 |
ENST00000270189.6 ENST00000352041.3 ENST00000220809.4 |
PLAT |
plasminogen activator, tissue |
| chr4_-_73935409 | 0.25 |
ENST00000507544.2 ENST00000295890.4 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
| chr17_+_16284104 | 0.25 |
ENST00000577958.1 ENST00000302182.3 ENST00000577640.1 |
UBB |
ubiquitin B |
| chr3_+_158787041 | 0.25 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr17_-_53809473 | 0.25 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr5_+_140718396 | 0.25 |
ENST00000394576.2 |
PCDHGA2 |
protocadherin gamma subfamily A, 2 |
| chr7_+_79765071 | 0.24 |
ENST00000457358.2 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr8_-_42065075 | 0.24 |
ENST00000429089.2 ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT |
plasminogen activator, tissue |
| chr11_+_114310164 | 0.24 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr2_-_151344172 | 0.23 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr6_-_33168391 | 0.23 |
ENST00000374685.4 ENST00000413614.2 ENST00000374680.3 |
RXRB |
retinoid X receptor, beta |
| chr19_-_47164386 | 0.23 |
ENST00000391916.2 ENST00000410105.2 |
DACT3 |
dishevelled-binding antagonist of beta-catenin 3 |
| chr11_+_28129795 | 0.22 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr17_+_35851570 | 0.22 |
ENST00000394386.1 |
DUSP14 |
dual specificity phosphatase 14 |
| chr10_-_126716459 | 0.22 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr14_+_74111578 | 0.21 |
ENST00000554113.1 ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1 |
dynein, axonemal, light chain 1 |
| chr12_-_118796910 | 0.21 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr3_-_49142178 | 0.20 |
ENST00000452739.1 ENST00000414533.1 ENST00000417025.1 |
QARS |
glutaminyl-tRNA synthetase |
| chr7_+_138943265 | 0.20 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chrX_+_107288197 | 0.20 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr1_-_111970353 | 0.20 |
ENST00000369732.3 |
OVGP1 |
oviductal glycoprotein 1, 120kDa |
| chr14_-_54955721 | 0.19 |
ENST00000554908.1 |
GMFB |
glia maturation factor, beta |
| chr16_-_72206034 | 0.19 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr13_+_49684445 | 0.19 |
ENST00000398316.3 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr2_+_170366203 | 0.19 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr18_+_616672 | 0.18 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr6_+_26087646 | 0.18 |
ENST00000309234.6 |
HFE |
hemochromatosis |
| chr5_+_68530697 | 0.18 |
ENST00000256443.3 ENST00000514676.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chr14_+_96722152 | 0.18 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr9_-_21351377 | 0.18 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr1_+_57320437 | 0.17 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr7_+_80275953 | 0.17 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr3_-_49142504 | 0.17 |
ENST00000306125.6 ENST00000420147.2 |
QARS |
glutaminyl-tRNA synthetase |
| chr1_+_15802594 | 0.17 |
ENST00000375910.3 |
CELA2B |
chymotrypsin-like elastase family, member 2B |
| chr8_-_120605194 | 0.17 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_+_129083772 | 0.17 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr4_+_154073469 | 0.16 |
ENST00000441616.1 |
TRIM2 |
tripartite motif containing 2 |
| chr4_+_154622652 | 0.16 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr20_+_30795664 | 0.16 |
ENST00000375749.3 ENST00000375730.3 ENST00000539210.1 |
POFUT1 |
protein O-fucosyltransferase 1 |
| chr11_+_61015594 | 0.16 |
ENST00000451616.2 |
PGA5 |
pepsinogen 5, group I (pepsinogen A) |
| chr10_+_102295616 | 0.16 |
ENST00000299163.6 |
HIF1AN |
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr7_-_143454789 | 0.16 |
ENST00000470691.2 |
CTAGE6 |
CTAGE family, member 6 |
| chrX_+_107288239 | 0.16 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr3_-_155394152 | 0.16 |
ENST00000494598.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr14_+_96722539 | 0.16 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr7_+_80267973 | 0.15 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_-_90049828 | 0.15 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_+_57232690 | 0.15 |
ENST00000262293.4 |
PRR11 |
proline rich 11 |
| chr8_+_38585704 | 0.15 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr12_-_81763184 | 0.15 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr5_-_138775177 | 0.15 |
ENST00000302060.5 |
DNAJC18 |
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_+_140165876 | 0.15 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr12_+_72079842 | 0.14 |
ENST00000266673.5 ENST00000550524.1 |
TMEM19 |
transmembrane protein 19 |
| chr3_+_119501557 | 0.14 |
ENST00000337940.4 |
NR1I2 |
nuclear receptor subfamily 1, group I, member 2 |
| chr8_-_145047688 | 0.14 |
ENST00000356346.3 |
PLEC |
plectin |
| chr2_+_113763031 | 0.14 |
ENST00000259211.6 |
IL36A |
interleukin 36, alpha |
| chr15_-_66797172 | 0.14 |
ENST00000569438.1 ENST00000569696.1 ENST00000307961.6 |
RPL4 |
ribosomal protein L4 |
| chr3_-_37216055 | 0.14 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr1_+_171283331 | 0.14 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chr8_+_107738240 | 0.14 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr6_-_116447283 | 0.13 |
ENST00000452729.1 ENST00000243222.4 |
COL10A1 |
collagen, type X, alpha 1 |
| chr19_+_8455077 | 0.12 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr12_-_101604185 | 0.12 |
ENST00000536262.2 |
SLC5A8 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chrX_-_99987088 | 0.12 |
ENST00000372981.1 ENST00000263033.5 |
SYTL4 |
synaptotagmin-like 4 |
| chr18_+_616711 | 0.12 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr3_-_155394099 | 0.12 |
ENST00000414191.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr2_-_179672142 | 0.11 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr19_+_44598534 | 0.11 |
ENST00000336976.6 |
ZNF224 |
zinc finger protein 224 |
| chr22_-_37213554 | 0.11 |
ENST00000443735.1 |
PVALB |
parvalbumin |
| chr5_-_131879205 | 0.11 |
ENST00000231454.1 |
IL5 |
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr10_+_57358750 | 0.11 |
ENST00000512524.2 |
MTRNR2L5 |
MT-RNR2-like 5 |
| chr8_+_12803176 | 0.10 |
ENST00000524591.2 |
KIAA1456 |
KIAA1456 |
| chr17_+_40996590 | 0.10 |
ENST00000253799.3 ENST00000452774.2 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
| chr8_+_22422749 | 0.10 |
ENST00000523900.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr5_+_115177178 | 0.09 |
ENST00000316788.7 |
AP3S1 |
adaptor-related protein complex 3, sigma 1 subunit |
| chr2_-_224467093 | 0.09 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr20_+_46130619 | 0.09 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr8_-_16035454 | 0.09 |
ENST00000355282.2 |
MSR1 |
macrophage scavenger receptor 1 |
| chr6_-_52774464 | 0.09 |
ENST00000370968.1 ENST00000211122.3 |
GSTA3 |
glutathione S-transferase alpha 3 |
| chr6_+_32938692 | 0.09 |
ENST00000443797.2 |
BRD2 |
bromodomain containing 2 |
| chr6_+_161123270 | 0.09 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr6_+_46761118 | 0.08 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr19_-_49496557 | 0.08 |
ENST00000323798.3 ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1 |
glycogen synthase 1 (muscle) |
| chr11_-_102826434 | 0.08 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chrX_-_122756660 | 0.08 |
ENST00000441692.1 |
THOC2 |
THO complex 2 |
| chr3_+_171561127 | 0.08 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr12_-_10324716 | 0.08 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chrX_+_18725758 | 0.07 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr14_+_22615942 | 0.07 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_140602904 | 0.07 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr8_-_7320974 | 0.06 |
ENST00000528943.1 ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B |
sperm associated antigen 11B |
| chr3_-_151047327 | 0.06 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr9_-_21305312 | 0.06 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr11_-_61687739 | 0.06 |
ENST00000531922.1 ENST00000301773.5 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr5_-_138210977 | 0.06 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr17_-_73663168 | 0.06 |
ENST00000578201.1 ENST00000423245.2 |
RECQL5 |
RecQ protein-like 5 |
| chr11_-_61129335 | 0.05 |
ENST00000545361.1 ENST00000539128.1 ENST00000546151.1 ENST00000447532.2 |
CYB561A3 |
cytochrome b561 family, member A3 |
| chr9_-_123239632 | 0.05 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr3_-_183273477 | 0.05 |
ENST00000341319.3 |
KLHL6 |
kelch-like family member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.7 | 2.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 2.2 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.4 | 2.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.4 | 4.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 1.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.4 | 2.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 2.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 1.6 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.3 | 1.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 3.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 1.6 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.3 | 2.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.9 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 1.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.2 | 5.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.2 | 4.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 0.8 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 0.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.2 | 0.9 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.5 | GO:1901660 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.8 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.7 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.6 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.3 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 4.7 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.1 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.0 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.5 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 1.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 2.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 8.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 2.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.0 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.6 | 4.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 2.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 1.6 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 2.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 2.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 7.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 4.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 6.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 4.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 3.3 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |