Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
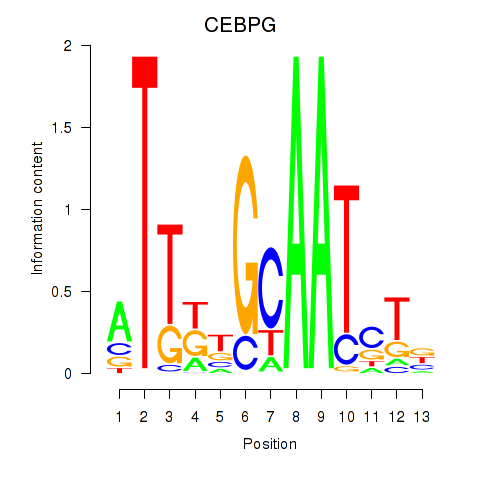
Results for CEBPG
Z-value: 1.06
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CEBPG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPG | hg19_v2_chr19_+_33865218_33865254 | 0.34 | 2.0e-01 | Click! |
Activity profile of CEBPG motif
Sorted Z-values of CEBPG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_186330712 | 5.15 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr3_-_148939835 | 4.84 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr4_+_74269956 | 4.72 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr9_+_117092149 | 4.52 |
ENST00000431067.2 ENST00000412657.1 |
ORM2 |
orosomucoid 2 |
| chr2_-_21266935 | 4.29 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr9_+_117085336 | 4.08 |
ENST00000259396.8 ENST00000538816.1 |
ORM1 |
orosomucoid 1 |
| chr19_-_6720686 | 3.73 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr13_-_46679185 | 3.54 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 3.51 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr8_+_97597148 | 3.40 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr6_+_31895254 | 3.36 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr17_-_64225508 | 3.31 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_11674213 | 3.30 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr6_+_31895467 | 2.97 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_+_31895480 | 2.88 |
ENST00000418949.2 ENST00000383177.3 ENST00000477310.1 |
C2 CFB |
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr4_+_184020398 | 2.54 |
ENST00000403733.3 ENST00000378925.3 |
WWC2 |
WW and C2 domain containing 2 |
| chr16_+_72088376 | 2.26 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chrX_+_46937745 | 2.21 |
ENST00000397180.1 ENST00000457380.1 ENST00000352078.4 |
RGN |
regucalcin |
| chr11_-_18258342 | 2.20 |
ENST00000278222.4 |
SAA4 |
serum amyloid A4, constitutive |
| chr3_-_49726486 | 2.04 |
ENST00000449682.2 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr17_-_42200958 | 1.96 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr17_-_42200996 | 1.79 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr2_+_102721023 | 1.60 |
ENST00000409589.1 ENST00000409329.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr3_-_194072019 | 1.52 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr18_+_33767473 | 1.49 |
ENST00000261326.5 |
MOCOS |
molybdenum cofactor sulfurase |
| chr17_-_79895154 | 1.29 |
ENST00000405481.4 ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr17_-_79895097 | 1.19 |
ENST00000402252.2 ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1 |
pyrroline-5-carboxylate reductase 1 |
| chr5_-_121413974 | 1.15 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr12_-_71551868 | 1.12 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr3_+_186435137 | 1.11 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr6_+_167704798 | 1.09 |
ENST00000230256.3 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr12_-_96390063 | 1.04 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr6_+_167704838 | 1.04 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr19_-_4540486 | 1.03 |
ENST00000306390.6 |
LRG1 |
leucine-rich alpha-2-glycoprotein 1 |
| chr2_-_216300784 | 0.99 |
ENST00000421182.1 ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1 |
fibronectin 1 |
| chr8_-_121824374 | 0.92 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chrX_+_49644470 | 0.92 |
ENST00000508866.2 |
USP27X |
ubiquitin specific peptidase 27, X-linked |
| chr20_+_361261 | 0.88 |
ENST00000217233.3 |
TRIB3 |
tribbles pseudokinase 3 |
| chr12_-_71551652 | 0.84 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr12_-_96390108 | 0.75 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr12_+_57522258 | 0.74 |
ENST00000553277.1 ENST00000243077.3 |
LRP1 |
low density lipoprotein receptor-related protein 1 |
| chr9_+_80912059 | 0.73 |
ENST00000347159.2 ENST00000376588.3 |
PSAT1 |
phosphoserine aminotransferase 1 |
| chr10_-_116444371 | 0.73 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr2_-_112614424 | 0.72 |
ENST00000427997.1 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr1_-_161277210 | 0.70 |
ENST00000491222.2 |
MPZ |
myelin protein zero |
| chr4_-_10023095 | 0.68 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr17_+_2699697 | 0.68 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chrX_-_73512411 | 0.66 |
ENST00000602576.1 ENST00000429124.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr22_-_31364187 | 0.65 |
ENST00000215862.4 ENST00000397641.3 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr6_-_135271260 | 0.63 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr21_-_35899113 | 0.60 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr17_-_61850894 | 0.59 |
ENST00000403162.3 ENST00000582252.1 ENST00000225726.5 |
CCDC47 |
coiled-coil domain containing 47 |
| chr3_+_73110810 | 0.58 |
ENST00000533473.1 |
EBLN2 |
endogenous Bornavirus-like nucleoprotein 2 |
| chrX_+_47082408 | 0.55 |
ENST00000518022.1 ENST00000276052.6 |
CDK16 |
cyclin-dependent kinase 16 |
| chr17_-_76124711 | 0.52 |
ENST00000306591.7 ENST00000590602.1 |
TMC6 |
transmembrane channel-like 6 |
| chr6_-_135271219 | 0.50 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr6_-_27799305 | 0.49 |
ENST00000357549.2 |
HIST1H4K |
histone cluster 1, H4k |
| chr20_-_48782639 | 0.45 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr21_+_34775181 | 0.45 |
ENST00000290219.6 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr1_+_52682052 | 0.44 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr1_+_245133062 | 0.44 |
ENST00000366523.1 |
EFCAB2 |
EF-hand calcium binding domain 2 |
| chr1_+_110162448 | 0.37 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr10_-_92681033 | 0.35 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr4_+_71226468 | 0.35 |
ENST00000226460.4 |
SMR3A |
submaxillary gland androgen regulated protein 3A |
| chr6_-_30710510 | 0.32 |
ENST00000376389.3 |
FLOT1 |
flotillin 1 |
| chr14_+_39703084 | 0.29 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr3_+_40498783 | 0.28 |
ENST00000338970.6 ENST00000396203.2 ENST00000416518.1 |
RPL14 |
ribosomal protein L14 |
| chr6_-_30710447 | 0.27 |
ENST00000456573.2 |
FLOT1 |
flotillin 1 |
| chr5_+_36152163 | 0.27 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr17_+_34900737 | 0.26 |
ENST00000304718.4 ENST00000485685.2 |
GGNBP2 |
gametogenetin binding protein 2 |
| chr4_-_111119804 | 0.26 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr2_-_89545079 | 0.25 |
ENST00000468494.1 |
IGKV2-30 |
immunoglobulin kappa variable 2-30 |
| chr15_-_34628951 | 0.25 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr3_+_186435065 | 0.25 |
ENST00000287611.2 ENST00000265023.4 |
KNG1 |
kininogen 1 |
| chr6_+_36238237 | 0.25 |
ENST00000457797.1 ENST00000394571.2 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
| chr11_-_72504637 | 0.24 |
ENST00000536377.1 ENST00000359373.5 |
STARD10 ARAP1 |
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr5_+_36152091 | 0.21 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr14_-_80677815 | 0.20 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr8_+_38585704 | 0.20 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_5248294 | 0.20 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr1_+_154377669 | 0.20 |
ENST00000368485.3 ENST00000344086.4 |
IL6R |
interleukin 6 receptor |
| chr2_-_183291741 | 0.18 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr12_-_13248598 | 0.17 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr2_+_226265364 | 0.16 |
ENST00000272907.6 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr11_+_60048053 | 0.15 |
ENST00000337908.4 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr12_-_114841703 | 0.14 |
ENST00000526441.1 |
TBX5 |
T-box 5 |
| chr12_-_13248562 | 0.14 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chrX_-_73512177 | 0.14 |
ENST00000603672.1 ENST00000418855.1 |
FTX |
FTX transcript, XIST regulator (non-protein coding) |
| chr14_-_50319758 | 0.14 |
ENST00000298310.5 |
NEMF |
nuclear export mediator factor |
| chr1_+_165864821 | 0.14 |
ENST00000470820.1 |
UCK2 |
uridine-cytidine kinase 2 |
| chr17_+_61851504 | 0.13 |
ENST00000359353.5 ENST00000389924.2 |
DDX42 |
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr14_+_96000930 | 0.13 |
ENST00000331334.4 |
GLRX5 |
glutaredoxin 5 |
| chr2_-_32390801 | 0.12 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr16_+_6069586 | 0.11 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_+_96722539 | 0.10 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr5_-_145562147 | 0.09 |
ENST00000545646.1 ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS |
leucyl-tRNA synthetase |
| chr11_+_59824127 | 0.09 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr4_+_71248795 | 0.09 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr17_+_61851157 | 0.08 |
ENST00000578681.1 ENST00000583590.1 |
DDX42 |
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr1_+_172389821 | 0.08 |
ENST00000367727.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr4_+_71600144 | 0.08 |
ENST00000502653.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr11_+_59824060 | 0.07 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr5_-_146258205 | 0.06 |
ENST00000394413.3 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_158901329 | 0.06 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr5_+_36152179 | 0.06 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr16_+_6069072 | 0.06 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr19_+_55417530 | 0.05 |
ENST00000350790.5 ENST00000338835.5 ENST00000357397.5 |
NCR1 |
natural cytotoxicity triggering receptor 1 |
| chr12_-_10605929 | 0.05 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr13_-_36788718 | 0.05 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr12_-_323689 | 0.04 |
ENST00000428720.1 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr11_+_60048129 | 0.04 |
ENST00000355131.3 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr21_-_37852359 | 0.04 |
ENST00000399137.1 ENST00000399135.1 |
CLDN14 |
claudin 14 |
| chr14_-_50319482 | 0.04 |
ENST00000546046.1 ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF |
nuclear export mediator factor |
| chr1_+_165864800 | 0.04 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr8_+_42552533 | 0.03 |
ENST00000289957.2 |
CHRNB3 |
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr19_-_36822595 | 0.03 |
ENST00000585356.1 ENST00000438368.2 ENST00000590622.1 |
LINC00665 |
long intergenic non-protein coding RNA 665 |
| chr19_+_55417499 | 0.03 |
ENST00000291890.4 ENST00000447255.1 ENST00000598576.1 ENST00000594765.1 |
NCR1 |
natural cytotoxicity triggering receptor 1 |
| chr14_-_80677970 | 0.01 |
ENST00000438257.4 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr12_-_8693539 | 0.01 |
ENST00000299663.3 |
CLEC4E |
C-type lectin domain family 4, member E |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.6 | 4.7 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.2 | 3.7 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 1.1 | 8.6 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.8 | 2.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.7 | 2.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.6 | 4.3 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.6 | 1.8 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 6.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 1.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.5 | 1.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.5 | 3.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 2.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 3.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.4 | 2.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.7 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.2 | 5.2 | GO:0030502 | pinocytosis(GO:0006907) negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 3.8 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 | 1.5 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 4.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.7 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 3.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.4 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.5 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.6 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.9 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.9 | GO:0071108 | protein K63-linked deubiquitination(GO:0070536) protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 1.4 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.7 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.0 | 0.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 1.8 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.9 | 3.8 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.7 | 3.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 4.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 1.5 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.3 | 4.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 8.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.7 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 3.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.9 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 4.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 2.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 4.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 4.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 3.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 5.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 5.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 4.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 3.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 20.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 14.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0005802 | trans-Golgi network(GO:0005802) |