Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
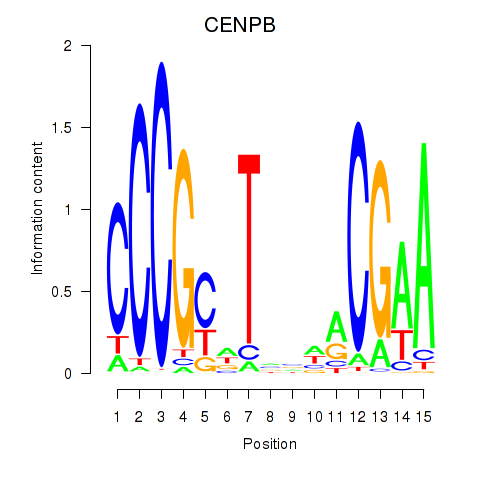
Results for CENPB
Z-value: 0.54
Transcription factors associated with CENPB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CENPB
|
ENSG00000125817.7 | CENPB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CENPB | hg19_v2_chr20_-_3767324_3767443 | -0.23 | 3.8e-01 | Click! |
Activity profile of CENPB motif
Sorted Z-values of CENPB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CENPB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 0.96 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chrY_+_2709906 | 0.89 |
ENST00000430575.1 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr11_-_61582579 | 0.83 |
ENST00000539419.1 ENST00000545245.1 ENST00000545405.1 ENST00000542506.1 |
FADS1 |
fatty acid desaturase 1 |
| chr6_+_30525051 | 0.54 |
ENST00000376557.3 |
PRR3 |
proline rich 3 |
| chr17_+_53342311 | 0.50 |
ENST00000226067.5 |
HLF |
hepatic leukemia factor |
| chr6_+_30524663 | 0.49 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr10_+_22610124 | 0.45 |
ENST00000376663.3 |
BMI1 |
BMI1 polycomb ring finger oncogene |
| chr10_+_30722866 | 0.44 |
ENST00000263056.1 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
| chr7_-_150038704 | 0.42 |
ENST00000466675.1 ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2 |
retinoic acid receptor responder (tazarotene induced) 2 |
| chr11_-_66115032 | 0.39 |
ENST00000311181.4 |
B3GNT1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr18_+_268148 | 0.35 |
ENST00000581677.1 |
RP11-705O1.8 |
RP11-705O1.8 |
| chr6_-_39197226 | 0.34 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr22_+_19467261 | 0.33 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chr9_-_130953731 | 0.32 |
ENST00000420484.1 ENST00000372954.1 ENST00000541172.1 ENST00000357558.5 ENST00000325721.8 ENST00000372938.5 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chr17_-_60142609 | 0.31 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr6_-_26124138 | 0.30 |
ENST00000314332.5 ENST00000396984.1 |
HIST1H2BC |
histone cluster 1, H2bc |
| chr7_-_99698338 | 0.30 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr2_-_152684977 | 0.29 |
ENST00000428992.2 ENST00000295087.8 |
ARL5A |
ADP-ribosylation factor-like 5A |
| chr17_-_72869086 | 0.28 |
ENST00000581530.1 ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR |
ferredoxin reductase |
| chr18_-_268019 | 0.28 |
ENST00000261600.6 |
THOC1 |
THO complex 1 |
| chr11_-_118927816 | 0.27 |
ENST00000534233.1 ENST00000532752.1 ENST00000525859.1 ENST00000404233.3 ENST00000532421.1 ENST00000543287.1 ENST00000527310.2 ENST00000529972.1 |
HYOU1 |
hypoxia up-regulated 1 |
| chr11_+_59522900 | 0.27 |
ENST00000529177.1 |
STX3 |
syntaxin 3 |
| chr2_+_73114489 | 0.27 |
ENST00000234454.5 |
SPR |
sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) |
| chr22_+_23412479 | 0.27 |
ENST00000248996.4 |
GNAZ |
guanine nucleotide binding protein (G protein), alpha z polypeptide |
| chr17_+_38296576 | 0.26 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr15_+_45879534 | 0.25 |
ENST00000564080.1 ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4 BLOC1S6 |
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr3_-_15469006 | 0.24 |
ENST00000443029.1 ENST00000383790.3 ENST00000383789.5 |
METTL6 |
methyltransferase like 6 |
| chr11_+_59522837 | 0.24 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr1_-_114355083 | 0.24 |
ENST00000261441.5 |
RSBN1 |
round spermatid basic protein 1 |
| chr12_+_110906169 | 0.23 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr3_+_173116225 | 0.23 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr14_+_50779071 | 0.22 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr19_-_45996465 | 0.22 |
ENST00000430715.2 |
RTN2 |
reticulon 2 |
| chr16_-_30569584 | 0.22 |
ENST00000252797.2 ENST00000568114.1 |
ZNF764 AC002310.13 |
zinc finger protein 764 Uncharacterized protein |
| chr6_+_26124373 | 0.21 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr17_-_72869140 | 0.21 |
ENST00000583917.1 ENST00000293195.5 ENST00000442102.2 |
FDXR |
ferredoxin reductase |
| chr19_+_34919257 | 0.21 |
ENST00000246548.4 ENST00000590048.2 |
UBA2 |
ubiquitin-like modifier activating enzyme 2 |
| chr2_-_103353277 | 0.21 |
ENST00000258436.5 |
MFSD9 |
major facilitator superfamily domain containing 9 |
| chr3_-_142166904 | 0.20 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr20_+_5107532 | 0.19 |
ENST00000450570.1 ENST00000379062.4 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr19_+_50180317 | 0.19 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr4_-_165305086 | 0.19 |
ENST00000507270.1 ENST00000514618.1 ENST00000503008.1 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_+_108487245 | 0.19 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr20_-_41818373 | 0.18 |
ENST00000373187.1 ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT |
protein tyrosine phosphatase, receptor type, T |
| chr9_-_99775862 | 0.18 |
ENST00000602917.1 ENST00000375223.4 |
HIATL2 |
hippocampus abundant transcript-like 2 |
| chr20_-_41818536 | 0.18 |
ENST00000373193.3 ENST00000373198.4 ENST00000373201.1 |
PTPRT |
protein tyrosine phosphatase, receptor type, T |
| chr12_-_110906027 | 0.18 |
ENST00000537466.2 ENST00000550974.1 ENST00000228827.3 |
GPN3 |
GPN-loop GTPase 3 |
| chr22_-_20104700 | 0.18 |
ENST00000439169.2 ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr11_+_64009072 | 0.17 |
ENST00000535135.1 ENST00000394540.3 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chrX_-_135962876 | 0.17 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr17_+_72427477 | 0.16 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr15_-_49447835 | 0.16 |
ENST00000388901.5 ENST00000299259.6 |
COPS2 |
COP9 signalosome subunit 2 |
| chr7_+_50348268 | 0.16 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr20_-_45061695 | 0.15 |
ENST00000445496.2 |
ELMO2 |
engulfment and cell motility 2 |
| chr6_-_166582107 | 0.15 |
ENST00000296946.2 ENST00000461348.2 ENST00000366871.3 |
T |
T, brachyury homolog (mouse) |
| chr20_+_306177 | 0.15 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr19_+_12075844 | 0.15 |
ENST00000592625.1 ENST00000586494.1 ENST00000343949.5 ENST00000545530.1 ENST00000358987.3 |
ZNF763 |
zinc finger protein 763 |
| chr20_-_55841662 | 0.15 |
ENST00000395863.3 ENST00000450594.2 |
BMP7 |
bone morphogenetic protein 7 |
| chr20_+_306221 | 0.15 |
ENST00000342665.2 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr14_-_45722605 | 0.15 |
ENST00000310806.4 |
MIS18BP1 |
MIS18 binding protein 1 |
| chr14_+_103058948 | 0.15 |
ENST00000262241.6 |
RCOR1 |
REST corepressor 1 |
| chr20_-_55841398 | 0.14 |
ENST00000395864.3 |
BMP7 |
bone morphogenetic protein 7 |
| chr16_-_66968265 | 0.13 |
ENST00000567511.1 ENST00000422424.2 |
FAM96B |
family with sequence similarity 96, member B |
| chr14_+_50779029 | 0.13 |
ENST00000245448.6 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr11_+_126152954 | 0.13 |
ENST00000392679.1 |
TIRAP |
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr1_-_167905225 | 0.13 |
ENST00000367846.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr22_+_39853258 | 0.12 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr16_-_30569801 | 0.12 |
ENST00000395091.2 |
ZNF764 |
zinc finger protein 764 |
| chr9_-_136039282 | 0.12 |
ENST00000372036.3 ENST00000372038.3 ENST00000540636.1 ENST00000372043.3 ENST00000542690.1 |
GBGT1 RALGDS |
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 ral guanine nucleotide dissociation stimulator |
| chr9_-_123342415 | 0.12 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr19_+_41882598 | 0.12 |
ENST00000447302.2 ENST00000544232.1 ENST00000542945.1 ENST00000540732.1 |
TMEM91 CTC-435M10.3 |
transmembrane protein 91 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial; Uncharacterized protein |
| chr1_-_151319283 | 0.11 |
ENST00000392746.3 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr11_+_134201768 | 0.11 |
ENST00000535456.2 ENST00000339772.7 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr2_-_136875712 | 0.11 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr16_-_19729502 | 0.11 |
ENST00000219837.7 |
KNOP1 |
lysine-rich nucleolar protein 1 |
| chr1_-_156390128 | 0.10 |
ENST00000368242.3 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr7_-_102257139 | 0.10 |
ENST00000521076.1 ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4 |
RAS p21 protein activator 4 |
| chr11_-_61197480 | 0.10 |
ENST00000439958.3 ENST00000394888.4 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
| chr19_-_663277 | 0.10 |
ENST00000292363.5 |
RNF126 |
ring finger protein 126 |
| chr9_-_127533582 | 0.10 |
ENST00000416460.2 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chr2_-_74757066 | 0.09 |
ENST00000377526.3 |
AUP1 |
ancient ubiquitous protein 1 |
| chr7_+_100303676 | 0.09 |
ENST00000303151.4 |
POP7 |
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr8_+_56685701 | 0.09 |
ENST00000260129.5 |
TGS1 |
trimethylguanosine synthase 1 |
| chr17_-_64188177 | 0.08 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr3_-_196669371 | 0.08 |
ENST00000427641.2 ENST00000321256.5 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
| chr2_-_10952832 | 0.08 |
ENST00000540494.1 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr16_-_66968055 | 0.07 |
ENST00000568572.1 |
FAM96B |
family with sequence similarity 96, member B |
| chr18_-_51750948 | 0.07 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr2_+_74757050 | 0.07 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr4_-_170533723 | 0.07 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr7_-_158937633 | 0.07 |
ENST00000262178.2 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
| chrX_+_147582130 | 0.07 |
ENST00000370460.2 ENST00000370457.5 |
AFF2 |
AF4/FMR2 family, member 2 |
| chr1_+_8021954 | 0.07 |
ENST00000377491.1 ENST00000377488.1 |
PARK7 |
parkinson protein 7 |
| chr3_+_182511266 | 0.07 |
ENST00000323116.5 ENST00000493826.1 |
ATP11B |
ATPase, class VI, type 11B |
| chr3_+_52232102 | 0.06 |
ENST00000469224.1 ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr16_-_20367584 | 0.06 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chr20_-_52790512 | 0.06 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr19_+_49588677 | 0.05 |
ENST00000598984.1 ENST00000598441.1 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
| chr7_-_102158157 | 0.05 |
ENST00000541662.1 ENST00000306682.6 ENST00000465829.1 |
RASA4B |
RAS p21 protein activator 4B |
| chr11_+_67033881 | 0.05 |
ENST00000308595.5 ENST00000526285.1 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
| chrX_+_151081351 | 0.04 |
ENST00000276344.2 |
MAGEA4 |
melanoma antigen family A, 4 |
| chr1_+_25664408 | 0.04 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr6_-_30524951 | 0.04 |
ENST00000376621.3 |
GNL1 |
guanine nucleotide binding protein-like 1 |
| chr1_+_151372010 | 0.03 |
ENST00000290541.6 |
PSMB4 |
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr19_+_50180409 | 0.03 |
ENST00000391851.4 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr2_-_241500447 | 0.03 |
ENST00000536462.1 ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr22_+_20105012 | 0.03 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr6_+_150070831 | 0.03 |
ENST00000367380.5 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr11_-_71791726 | 0.03 |
ENST00000393695.3 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
| chr3_+_15468862 | 0.02 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr9_-_139096955 | 0.02 |
ENST00000371748.5 |
LHX3 |
LIM homeobox 3 |
| chr1_-_51810778 | 0.02 |
ENST00000413473.2 ENST00000401051.3 ENST00000527205.1 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr11_+_89764274 | 0.02 |
ENST00000448984.1 ENST00000432771.1 |
TRIM49C |
tripartite motif containing 49C |
| chr10_-_14996070 | 0.02 |
ENST00000378258.1 ENST00000453695.2 ENST00000378246.2 |
DCLRE1C |
DNA cross-link repair 1C |
| chr15_-_49447771 | 0.02 |
ENST00000558843.1 ENST00000542928.1 ENST00000561248.1 |
COPS2 |
COP9 signalosome subunit 2 |
| chr15_+_45879321 | 0.02 |
ENST00000220531.3 ENST00000567461.1 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr10_-_135187193 | 0.02 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr11_+_82612740 | 0.02 |
ENST00000524921.1 ENST00000528759.1 ENST00000525361.1 ENST00000430323.2 ENST00000533655.1 ENST00000532764.1 ENST00000532589.1 ENST00000525388.1 |
C11orf82 |
chromosome 11 open reading frame 82 |
| chr6_+_150070857 | 0.01 |
ENST00000544496.1 |
PCMT1 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr19_+_50180507 | 0.01 |
ENST00000454376.2 ENST00000524771.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr11_-_82612727 | 0.01 |
ENST00000531128.1 ENST00000535099.1 ENST00000527444.1 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.8 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.3 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:2000343 | positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.3 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 1.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 0.5 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.5 | GO:0050544 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |