Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
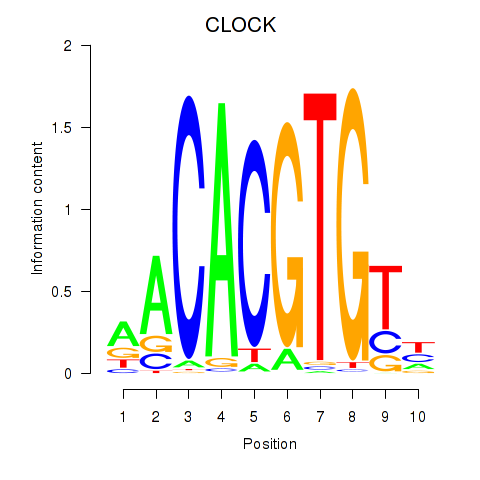
Results for CLOCK
Z-value: 0.75
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | CLOCK |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106322288 | 2.19 |
ENST00000390559.2 |
IGHM |
immunoglobulin heavy constant mu |
| chr12_-_21810726 | 1.78 |
ENST00000396076.1 |
LDHB |
lactate dehydrogenase B |
| chr12_-_21810765 | 1.77 |
ENST00000450584.1 ENST00000350669.1 |
LDHB |
lactate dehydrogenase B |
| chr3_-_127541679 | 1.65 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr4_+_41258786 | 1.60 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_-_127441406 | 1.40 |
ENST00000487473.1 ENST00000484451.1 |
MGLL |
monoglyceride lipase |
| chr12_+_81110684 | 1.38 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr11_-_73694346 | 1.20 |
ENST00000310473.3 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr3_-_127542051 | 1.12 |
ENST00000398104.1 |
MGLL |
monoglyceride lipase |
| chr14_+_61789382 | 1.10 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr9_-_35691017 | 1.06 |
ENST00000378292.3 |
TPM2 |
tropomyosin 2 (beta) |
| chr9_+_112542591 | 1.01 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr21_+_27011584 | 1.01 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr9_+_112542572 | 0.99 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr4_-_111563076 | 0.95 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr10_-_75415825 | 0.94 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr11_-_64014379 | 0.85 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_-_150537279 | 0.85 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr8_+_28196157 | 0.77 |
ENST00000522209.1 |
PNOC |
prepronociceptin |
| chr22_+_18593446 | 0.76 |
ENST00000316027.6 |
TUBA8 |
tubulin, alpha 8 |
| chr10_-_126849588 | 0.75 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr19_-_46974664 | 0.75 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr19_-_46974741 | 0.75 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr7_-_94285402 | 0.73 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr11_-_122931881 | 0.73 |
ENST00000526110.1 ENST00000227378.3 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr6_-_32636145 | 0.71 |
ENST00000399084.1 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr11_-_122932730 | 0.71 |
ENST00000532182.1 ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr16_+_89238149 | 0.68 |
ENST00000289746.2 |
CDH15 |
cadherin 15, type 1, M-cadherin (myotubule) |
| chr21_-_46330545 | 0.68 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_-_111312622 | 0.66 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr7_+_99816859 | 0.66 |
ENST00000317271.2 |
PVRIG |
poliovirus receptor related immunoglobulin domain containing |
| chr19_+_35168633 | 0.66 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr12_-_25102252 | 0.64 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr3_+_133292574 | 0.64 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr12_-_49318715 | 0.63 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr16_+_56965960 | 0.63 |
ENST00000439977.2 ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr20_+_44441215 | 0.62 |
ENST00000356455.4 ENST00000405520.1 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr16_+_6533380 | 0.61 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr20_+_44441304 | 0.61 |
ENST00000352551.5 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr17_+_30813576 | 0.60 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_-_159894319 | 0.59 |
ENST00000320307.4 |
TAGLN2 |
transgelin 2 |
| chr2_+_233527443 | 0.58 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr1_-_9129735 | 0.55 |
ENST00000377424.4 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr11_-_47470591 | 0.55 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr3_+_112280857 | 0.53 |
ENST00000492406.1 ENST00000468642.1 |
SLC35A5 |
solute carrier family 35, member A5 |
| chr12_-_25101920 | 0.53 |
ENST00000539780.1 ENST00000546285.1 ENST00000342945.5 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr15_+_81475047 | 0.52 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr1_+_21880560 | 0.50 |
ENST00000425315.2 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr11_-_47470703 | 0.49 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr7_+_100464760 | 0.48 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr5_+_155753745 | 0.48 |
ENST00000435422.3 ENST00000337851.4 ENST00000447401.1 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
| chr1_+_182992545 | 0.47 |
ENST00000258341.4 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
| chr13_-_95953589 | 0.47 |
ENST00000538287.1 ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr1_-_154909329 | 0.47 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr20_+_44441271 | 0.47 |
ENST00000335046.3 ENST00000243893.6 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr1_-_9129598 | 0.46 |
ENST00000535586.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr5_-_138725594 | 0.46 |
ENST00000302125.8 |
MZB1 |
marginal zone B and B1 cell-specific protein |
| chr5_-_138725560 | 0.46 |
ENST00000412103.2 ENST00000457570.2 |
MZB1 |
marginal zone B and B1 cell-specific protein |
| chr9_-_75567962 | 0.46 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr18_+_33877654 | 0.46 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr8_+_38614807 | 0.45 |
ENST00000330691.6 ENST00000348567.4 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_47470682 | 0.44 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr17_-_62009621 | 0.43 |
ENST00000349817.2 ENST00000392795.3 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
| chr1_-_9129895 | 0.43 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr14_-_91884150 | 0.43 |
ENST00000553403.1 |
CCDC88C |
coiled-coil domain containing 88C |
| chr11_-_66336060 | 0.43 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr4_-_76439483 | 0.43 |
ENST00000380840.2 ENST00000513257.1 ENST00000507014.1 |
RCHY1 |
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr6_-_32811771 | 0.42 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_62009702 | 0.42 |
ENST00000006750.3 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
| chr18_-_45457478 | 0.41 |
ENST00000402690.2 ENST00000356825.4 |
SMAD2 |
SMAD family member 2 |
| chr21_-_27542972 | 0.40 |
ENST00000346798.3 ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP |
amyloid beta (A4) precursor protein |
| chrX_+_11777671 | 0.40 |
ENST00000380693.3 ENST00000380692.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr3_+_50284321 | 0.40 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chrX_+_51636629 | 0.39 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr4_+_40198527 | 0.39 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr19_+_1071203 | 0.39 |
ENST00000543365.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr5_-_176900610 | 0.37 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chr4_+_169842707 | 0.37 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_-_9129631 | 0.37 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr17_+_75084717 | 0.36 |
ENST00000561721.2 ENST00000589827.1 ENST00000392476.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr4_-_103266626 | 0.36 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr16_+_84853580 | 0.35 |
ENST00000262424.5 ENST00000566151.1 ENST00000567845.1 ENST00000564567.1 ENST00000569090.1 |
CRISPLD2 |
cysteine-rich secretory protein LCCL domain containing 2 |
| chr5_+_110074685 | 0.35 |
ENST00000355943.3 ENST00000447245.2 |
SLC25A46 |
solute carrier family 25, member 46 |
| chr13_+_27998681 | 0.35 |
ENST00000381140.4 |
GTF3A |
general transcription factor IIIA |
| chr8_-_95908902 | 0.34 |
ENST00000520509.1 |
CCNE2 |
cyclin E2 |
| chrX_+_54556633 | 0.34 |
ENST00000336470.4 ENST00000360845.2 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr1_-_26232951 | 0.34 |
ENST00000426559.2 ENST00000455785.2 |
STMN1 |
stathmin 1 |
| chr3_+_133293278 | 0.34 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr2_+_217498105 | 0.33 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr6_-_154677900 | 0.33 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr1_+_84944926 | 0.33 |
ENST00000370656.1 ENST00000370654.5 |
RPF1 |
ribosome production factor 1 homolog (S. cerevisiae) |
| chr11_-_125366089 | 0.33 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr6_-_144385698 | 0.32 |
ENST00000444202.1 ENST00000437412.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr5_-_93447333 | 0.32 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr15_-_64126084 | 0.32 |
ENST00000560316.1 ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chrX_-_51239425 | 0.32 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr10_+_180405 | 0.31 |
ENST00000439456.1 ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr4_+_75023816 | 0.31 |
ENST00000395759.2 ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr5_+_152870106 | 0.31 |
ENST00000285900.5 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
| chr9_-_34620440 | 0.31 |
ENST00000421919.1 ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3 |
dynactin 3 (p22) |
| chr22_-_42343117 | 0.31 |
ENST00000407253.3 ENST00000215980.5 |
CENPM |
centromere protein M |
| chr2_-_11810284 | 0.30 |
ENST00000306928.5 |
NTSR2 |
neurotensin receptor 2 |
| chr5_-_102455801 | 0.30 |
ENST00000508629.1 ENST00000399004.2 |
GIN1 |
gypsy retrotransposon integrase 1 |
| chr4_-_103266355 | 0.30 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr19_+_21688366 | 0.29 |
ENST00000358491.4 ENST00000597078.1 |
ZNF429 |
zinc finger protein 429 |
| chr4_+_2814011 | 0.29 |
ENST00000502260.1 ENST00000435136.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr10_-_16859361 | 0.28 |
ENST00000377921.3 |
RSU1 |
Ras suppressor protein 1 |
| chr2_-_220118631 | 0.28 |
ENST00000248437.4 |
TUBA4A |
tubulin, alpha 4a |
| chr18_+_72163536 | 0.28 |
ENST00000579847.1 ENST00000583203.1 ENST00000581513.1 ENST00000577600.1 ENST00000579583.1 ENST00000584613.1 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr16_-_67194201 | 0.27 |
ENST00000345057.4 |
TRADD |
TNFRSF1A-associated via death domain |
| chr14_+_90422239 | 0.27 |
ENST00000393452.3 ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1 |
tyrosyl-DNA phosphodiesterase 1 |
| chr13_-_30424821 | 0.27 |
ENST00000380680.4 |
UBL3 |
ubiquitin-like 3 |
| chr10_-_16859442 | 0.27 |
ENST00000602389.1 ENST00000345264.5 |
RSU1 |
Ras suppressor protein 1 |
| chr17_-_7590745 | 0.27 |
ENST00000514944.1 ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53 |
tumor protein p53 |
| chr4_-_5891918 | 0.27 |
ENST00000512574.1 |
CRMP1 |
collapsin response mediator protein 1 |
| chr10_-_50747064 | 0.27 |
ENST00000355832.5 ENST00000603152.1 ENST00000447839.2 |
ERCC6 PGBD3 ERCC6-PGBD3 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr12_-_76477707 | 0.26 |
ENST00000551992.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr4_-_76439596 | 0.26 |
ENST00000451788.1 ENST00000512706.1 |
RCHY1 |
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr6_+_44191507 | 0.26 |
ENST00000371724.1 ENST00000371713.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_+_106809406 | 0.26 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr18_+_72163443 | 0.26 |
ENST00000324262.4 ENST00000580672.1 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr6_+_44191290 | 0.25 |
ENST00000371755.3 ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr20_+_57430162 | 0.25 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr6_+_44187334 | 0.25 |
ENST00000313248.7 ENST00000427851.2 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr13_-_36920872 | 0.25 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr4_+_17578815 | 0.25 |
ENST00000226299.4 |
LAP3 |
leucine aminopeptidase 3 |
| chr17_-_17942473 | 0.24 |
ENST00000585101.1 ENST00000474627.3 ENST00000444058.1 |
ATPAF2 |
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr22_+_22681656 | 0.24 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr17_+_42429493 | 0.24 |
ENST00000586242.1 |
GRN |
granulin |
| chr13_-_36920615 | 0.24 |
ENST00000494062.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr6_-_153304697 | 0.23 |
ENST00000367241.3 |
FBXO5 |
F-box protein 5 |
| chr15_+_74466012 | 0.23 |
ENST00000249842.3 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
| chr5_+_95066823 | 0.23 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr14_-_91526922 | 0.23 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr1_-_212873267 | 0.22 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr1_+_12123414 | 0.22 |
ENST00000263932.2 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
| chr17_+_17942684 | 0.22 |
ENST00000376345.3 |
GID4 |
GID complex subunit 4 |
| chr13_+_51483814 | 0.22 |
ENST00000336617.3 ENST00000422660.1 |
RNASEH2B |
ribonuclease H2, subunit B |
| chr4_-_103266219 | 0.21 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr6_-_153304148 | 0.21 |
ENST00000229758.3 |
FBXO5 |
F-box protein 5 |
| chr4_+_17579110 | 0.21 |
ENST00000606142.1 |
LAP3 |
leucine aminopeptidase 3 |
| chr6_+_292459 | 0.21 |
ENST00000419235.2 ENST00000605035.1 ENST00000605863.1 |
DUSP22 |
dual specificity phosphatase 22 |
| chr1_-_78149041 | 0.21 |
ENST00000414381.1 ENST00000370798.1 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
| chr11_+_66406149 | 0.21 |
ENST00000578778.1 ENST00000483858.1 ENST00000398692.4 ENST00000510173.2 ENST00000506523.2 ENST00000530235.1 ENST00000532968.1 |
RBM4 |
RNA binding motif protein 4 |
| chr2_-_231084820 | 0.21 |
ENST00000258382.5 ENST00000338556.3 |
SP110 |
SP110 nuclear body protein |
| chr11_+_66406088 | 0.20 |
ENST00000310092.7 ENST00000396053.4 ENST00000408993.2 |
RBM4 |
RNA binding motif protein 4 |
| chr9_-_6015607 | 0.20 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr14_-_23876801 | 0.20 |
ENST00000356287.3 |
MYH6 |
myosin, heavy chain 6, cardiac muscle, alpha |
| chr1_-_26233423 | 0.20 |
ENST00000357865.2 |
STMN1 |
stathmin 1 |
| chr9_+_103235365 | 0.20 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_+_214776516 | 0.20 |
ENST00000366955.3 |
CENPF |
centromere protein F, 350/400kDa |
| chr17_-_62084241 | 0.20 |
ENST00000449662.2 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr10_-_6019552 | 0.19 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr6_+_44187242 | 0.19 |
ENST00000393844.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr21_-_46237883 | 0.19 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr11_+_9595180 | 0.19 |
ENST00000450114.2 |
WEE1 |
WEE1 G2 checkpoint kinase |
| chr1_+_38478378 | 0.19 |
ENST00000373014.4 |
UTP11L |
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr21_-_45079341 | 0.19 |
ENST00000443485.1 ENST00000291560.2 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr15_+_68924327 | 0.18 |
ENST00000543950.1 |
CORO2B |
coronin, actin binding protein, 2B |
| chr6_+_7727030 | 0.18 |
ENST00000283147.6 |
BMP6 |
bone morphogenetic protein 6 |
| chr1_-_156710859 | 0.18 |
ENST00000361531.2 ENST00000412846.1 |
MRPL24 |
mitochondrial ribosomal protein L24 |
| chr11_-_64511789 | 0.18 |
ENST00000419843.1 ENST00000394430.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr12_+_6833237 | 0.18 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr1_-_156710916 | 0.18 |
ENST00000368211.4 |
MRPL24 |
mitochondrial ribosomal protein L24 |
| chr17_+_46970127 | 0.18 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr21_+_38071430 | 0.18 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr17_+_46970134 | 0.18 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chrX_+_24711997 | 0.18 |
ENST00000379068.3 ENST00000379059.3 |
POLA1 |
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr6_-_80657292 | 0.18 |
ENST00000369816.4 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
| chr2_-_10587897 | 0.17 |
ENST00000405333.1 ENST00000443218.1 |
ODC1 |
ornithine decarboxylase 1 |
| chr21_-_46238034 | 0.17 |
ENST00000332859.6 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr19_+_50180317 | 0.17 |
ENST00000534465.1 |
PRMT1 |
protein arginine methyltransferase 1 |
| chr17_+_46970178 | 0.17 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_+_6833437 | 0.17 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr3_-_183543301 | 0.17 |
ENST00000318631.3 ENST00000431348.1 |
MAP6D1 |
MAP6 domain containing 1 |
| chr19_+_1067271 | 0.17 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr1_+_32479430 | 0.17 |
ENST00000327300.7 ENST00000492989.1 |
KHDRBS1 |
KH domain containing, RNA binding, signal transduction associated 1 |
| chr9_+_117373486 | 0.17 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr2_-_231084617 | 0.16 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr5_+_172571445 | 0.16 |
ENST00000231668.9 ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1 |
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr1_-_241520525 | 0.16 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr4_-_2264015 | 0.16 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr16_+_81272287 | 0.16 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr13_-_44735393 | 0.16 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr3_-_112280709 | 0.16 |
ENST00000402314.2 ENST00000283290.5 ENST00000492886.1 |
ATG3 |
autophagy related 3 |
| chr2_-_179315490 | 0.15 |
ENST00000487082.1 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr5_+_172386517 | 0.15 |
ENST00000519522.1 |
RPL26L1 |
ribosomal protein L26-like 1 |
| chr1_-_36107445 | 0.15 |
ENST00000373237.3 |
PSMB2 |
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr14_+_96722152 | 0.15 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr1_+_29213678 | 0.15 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr12_+_52695617 | 0.15 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr11_+_71938925 | 0.15 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr6_-_99873145 | 0.15 |
ENST00000369239.5 ENST00000438806.1 |
PNISR |
PNN-interacting serine/arginine-rich protein |
| chr8_-_103424916 | 0.15 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr21_-_46237959 | 0.15 |
ENST00000397898.3 ENST00000411651.2 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr2_+_207024306 | 0.14 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_241500447 | 0.14 |
ENST00000536462.1 ENST00000405002.1 ENST00000441168.1 ENST00000403283.1 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr2_-_179315453 | 0.14 |
ENST00000432031.2 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr1_+_63833261 | 0.14 |
ENST00000371108.4 |
ALG6 |
ALG6, alpha-1,3-glucosyltransferase |
| chr1_+_110163709 | 0.14 |
ENST00000369840.2 ENST00000527846.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.8 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 1.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.5 | 1.5 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.3 | 1.0 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 1.4 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.3 | 1.8 | GO:0032445 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 3.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.7 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 0.6 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 1.2 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.9 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.4 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.1 | 1.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 1.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.3 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.2 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.1 | 0.4 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.3 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.2 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 2.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.3 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.5 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.6 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.4 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0006568 | tryptophan metabolic process(GO:0006568) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0044266 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0007129 | synapsis(GO:0007129) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.2 | 1.9 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.5 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.4 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 0.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 2.2 | GO:0042571 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034719 | SMN-Sm protein complex(GO:0034719) Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.8 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 3.9 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.3 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 1.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.3 | 4.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.8 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 2.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 1.2 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.5 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 1.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.5 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.3 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.3 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.8 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.7 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.3 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.4 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |