Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
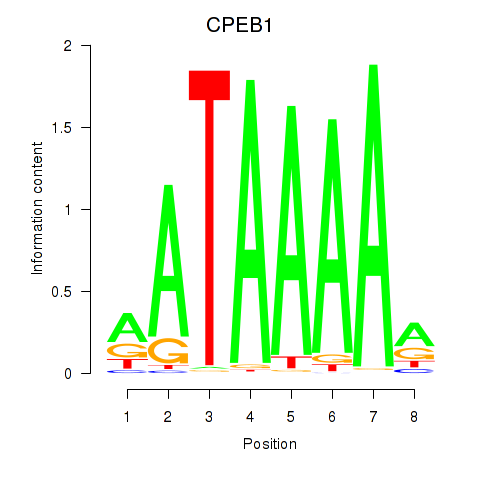
Results for CPEB1
Z-value: 1.28
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | CPEB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83316087_83316137 | 0.45 | 8.1e-02 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_151344172 | 7.05 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr2_-_190044480 | 4.49 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr14_-_75079026 | 4.26 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
| chr7_+_100464760 | 3.57 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chrX_+_135278908 | 3.27 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr10_+_17271266 | 3.11 |
ENST00000224237.5 |
VIM |
vimentin |
| chrX_+_135279179 | 2.88 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr3_+_158991025 | 2.81 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chrX_+_100805496 | 2.49 |
ENST00000372829.3 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
| chr2_-_175711133 | 2.45 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_-_27205136 | 2.38 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr4_+_174089904 | 2.28 |
ENST00000265000.4 |
GALNT7 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr17_+_45286706 | 2.21 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr3_+_159570722 | 2.14 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chrX_-_3264682 | 2.01 |
ENST00000217939.6 |
MXRA5 |
matrix-remodelling associated 5 |
| chr11_+_35160709 | 1.88 |
ENST00000415148.2 ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44 |
CD44 molecule (Indian blood group) |
| chr12_-_52967600 | 1.86 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr3_+_158787041 | 1.85 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr17_+_45286387 | 1.83 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr8_+_70378852 | 1.81 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr2_-_190927447 | 1.80 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr18_-_53177984 | 1.78 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr18_-_52989217 | 1.78 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr7_+_134551583 | 1.69 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr10_-_126847276 | 1.67 |
ENST00000531469.1 |
CTBP2 |
C-terminal binding protein 2 |
| chr18_+_61554932 | 1.67 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr12_-_52845910 | 1.65 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr12_-_91539918 | 1.64 |
ENST00000548218.1 |
DCN |
decorin |
| chr12_+_54422142 | 1.62 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr12_-_50616382 | 1.60 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr6_-_41909191 | 1.56 |
ENST00000512426.1 ENST00000372987.4 |
CCND3 |
cyclin D3 |
| chr18_-_53070913 | 1.54 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr11_-_27723158 | 1.51 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr8_-_108510224 | 1.51 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr11_-_5276008 | 1.51 |
ENST00000336906.4 |
HBG2 |
hemoglobin, gamma G |
| chr1_+_209848749 | 1.47 |
ENST00000367029.4 |
G0S2 |
G0/G1switch 2 |
| chr12_+_4385230 | 1.36 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr5_+_36608422 | 1.36 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_-_145277640 | 1.35 |
ENST00000539609.3 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_91546926 | 1.32 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_145277569 | 1.31 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr12_-_91572278 | 1.28 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr15_-_60690163 | 1.27 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr3_+_69928256 | 1.24 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr18_-_53253112 | 1.21 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr18_-_53253323 | 1.20 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr2_+_170366203 | 1.15 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr1_-_103574024 | 1.12 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr6_-_27114577 | 1.10 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr13_+_102142296 | 1.08 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_-_111563076 | 1.07 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr8_+_77593474 | 1.04 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr10_-_104262460 | 1.02 |
ENST00000446605.2 ENST00000369905.4 ENST00000545684.1 |
ACTR1A |
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr13_+_73632897 | 1.02 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr10_-_104262426 | 1.02 |
ENST00000487599.1 |
ACTR1A |
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr13_-_67802549 | 1.02 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr2_+_173292390 | 0.98 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr3_+_99357319 | 0.97 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr12_+_79258547 | 0.94 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chrX_-_48931648 | 0.94 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr3_-_141747439 | 0.93 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_173292280 | 0.92 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr2_-_211168332 | 0.92 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr12_+_79258444 | 0.92 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr5_+_32788945 | 0.91 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr4_+_74347400 | 0.88 |
ENST00000226355.3 |
AFM |
afamin |
| chr17_+_32582293 | 0.88 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr15_-_56209306 | 0.87 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr3_+_69788576 | 0.85 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr22_+_29876197 | 0.85 |
ENST00000310624.6 |
NEFH |
neurofilament, heavy polypeptide |
| chr11_+_12766583 | 0.84 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr4_-_52904425 | 0.84 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chrX_-_114252193 | 0.84 |
ENST00000243213.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chr11_-_8832182 | 0.84 |
ENST00000527510.1 ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5 |
suppression of tumorigenicity 5 |
| chr11_+_69924397 | 0.83 |
ENST00000355303.5 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr1_+_84630645 | 0.83 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_-_33702078 | 0.83 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr1_+_164528866 | 0.83 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr6_+_12290586 | 0.80 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr3_-_18480260 | 0.80 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr3_+_101546827 | 0.79 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr1_-_116311402 | 0.78 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr9_-_21995300 | 0.78 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_-_16870704 | 0.77 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr1_+_163039143 | 0.76 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr11_-_9025541 | 0.76 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr8_+_77593448 | 0.75 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr7_-_83824169 | 0.75 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr11_+_69924639 | 0.74 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr18_-_52989525 | 0.74 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr4_+_74735102 | 0.73 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr6_-_46293378 | 0.73 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr1_-_201391149 | 0.72 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr9_-_21995249 | 0.72 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr1_-_200379180 | 0.72 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr1_-_156675368 | 0.71 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr12_-_49412588 | 0.70 |
ENST00000547082.1 ENST00000395170.3 |
PRKAG1 |
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr1_+_84630053 | 0.70 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_39553844 | 0.69 |
ENST00000251645.2 |
KRT31 |
keratin 31 |
| chr3_-_37216055 | 0.69 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_49412541 | 0.69 |
ENST00000547306.1 ENST00000548857.1 ENST00000551696.1 ENST00000316299.5 |
PRKAG1 |
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr18_+_34124507 | 0.69 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr11_+_20385666 | 0.67 |
ENST00000532081.1 ENST00000531058.1 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr3_-_196910721 | 0.66 |
ENST00000443183.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr3_-_196911002 | 0.66 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr12_-_71148413 | 0.66 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr9_-_23825956 | 0.65 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr1_-_201390846 | 0.65 |
ENST00000367312.1 ENST00000555340.2 ENST00000361379.4 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr7_-_81399438 | 0.65 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399329 | 0.64 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_27100811 | 0.64 |
ENST00000359193.2 |
HIST1H2AG |
histone cluster 1, H2ag |
| chr6_+_39760783 | 0.64 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr4_-_102268484 | 0.64 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_29027696 | 0.64 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr2_-_55277654 | 0.63 |
ENST00000337526.6 ENST00000317610.7 ENST00000357732.4 |
RTN4 |
reticulon 4 |
| chr14_-_61748550 | 0.62 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr9_+_82186872 | 0.62 |
ENST00000376544.3 ENST00000376520.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr7_+_73442487 | 0.62 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr4_-_102268628 | 0.61 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_73442422 | 0.61 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr11_-_102651343 | 0.60 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr10_-_105845674 | 0.60 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr12_-_52828147 | 0.59 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr4_-_111119804 | 0.59 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chrX_-_15619076 | 0.58 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr11_+_34642656 | 0.58 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr6_-_31697977 | 0.57 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_13165457 | 0.56 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr2_+_88047606 | 0.56 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr17_-_10560619 | 0.56 |
ENST00000583535.1 |
MYH3 |
myosin, heavy chain 3, skeletal muscle, embryonic |
| chrX_-_77395186 | 0.55 |
ENST00000341864.5 |
TAF9B |
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr12_-_49412525 | 0.55 |
ENST00000551121.1 ENST00000552212.1 ENST00000548605.1 ENST00000548950.1 ENST00000547125.1 |
PRKAG1 |
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr7_-_27219849 | 0.55 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr11_-_57194948 | 0.53 |
ENST00000533235.1 ENST00000526621.1 ENST00000352187.1 |
SLC43A3 |
solute carrier family 43, member 3 |
| chr12_-_118797475 | 0.53 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr3_-_141747459 | 0.52 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_+_38211777 | 0.52 |
ENST00000039007.4 |
OTC |
ornithine carbamoyltransferase |
| chr7_-_50860565 | 0.51 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr10_-_62493223 | 0.51 |
ENST00000373827.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chrX_+_106871713 | 0.51 |
ENST00000372435.4 ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1 |
phosphoribosyl pyrophosphate synthetase 1 |
| chr3_+_44690211 | 0.51 |
ENST00000396056.2 ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35 |
zinc finger protein 35 |
| chr4_-_102267953 | 0.51 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr22_+_41258250 | 0.51 |
ENST00000544094.1 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr9_+_116263778 | 0.50 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr6_-_27100529 | 0.50 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chr7_+_73442457 | 0.50 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr5_+_140071178 | 0.50 |
ENST00000508522.1 ENST00000448069.2 |
HARS2 |
histidyl-tRNA synthetase 2, mitochondrial |
| chr17_-_39538550 | 0.49 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr12_+_28410128 | 0.49 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr8_+_85095553 | 0.49 |
ENST00000521268.1 |
RALYL |
RALY RNA binding protein-like |
| chr18_+_47088401 | 0.49 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr12_+_14518598 | 0.48 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr4_-_74904398 | 0.48 |
ENST00000296026.4 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
| chr11_-_62473706 | 0.47 |
ENST00000403550.1 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_-_10452929 | 0.47 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr1_-_152297679 | 0.46 |
ENST00000368799.1 |
FLG |
filaggrin |
| chr1_+_26798955 | 0.46 |
ENST00000361427.5 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
| chr5_-_82969405 | 0.46 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr11_-_62473776 | 0.45 |
ENST00000278893.7 ENST00000407022.3 ENST00000421906.1 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_21927736 | 0.45 |
ENST00000240662.2 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr8_-_86253888 | 0.45 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr1_-_200379129 | 0.44 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr13_-_103719196 | 0.44 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr8_-_61193947 | 0.43 |
ENST00000317995.4 |
CA8 |
carbonic anhydrase VIII |
| chr14_-_92413353 | 0.43 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr9_-_23826298 | 0.43 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr8_-_93029865 | 0.43 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_+_22040620 | 0.42 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr7_-_81399411 | 0.42 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr8_+_9953061 | 0.42 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr8_-_95274536 | 0.41 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr1_-_156675535 | 0.41 |
ENST00000368221.1 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr8_+_104831472 | 0.41 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr8_+_104892639 | 0.41 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_-_4852332 | 0.41 |
ENST00000572383.1 |
PFN1 |
profilin 1 |
| chr4_-_48782259 | 0.40 |
ENST00000507711.1 ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL |
FRY-like |
| chr18_+_3252206 | 0.39 |
ENST00000578562.2 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_+_169757750 | 0.38 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr2_-_87248975 | 0.38 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr5_+_162932554 | 0.38 |
ENST00000321757.6 ENST00000421814.2 ENST00000518095.1 |
MAT2B |
methionine adenosyltransferase II, beta |
| chr11_+_1940925 | 0.38 |
ENST00000453458.1 ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr18_+_3252265 | 0.37 |
ENST00000580887.1 ENST00000536605.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr6_-_161085291 | 0.37 |
ENST00000316300.5 |
LPA |
lipoprotein, Lp(a) |
| chr8_+_9953214 | 0.37 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr1_+_224301787 | 0.37 |
ENST00000366862.5 ENST00000424254.2 |
FBXO28 |
F-box protein 28 |
| chr1_-_200379104 | 0.36 |
ENST00000367352.3 |
ZNF281 |
zinc finger protein 281 |
| chr3_-_69129501 | 0.36 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
| chr5_-_140070897 | 0.36 |
ENST00000448240.1 ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS |
histidyl-tRNA synthetase |
| chr21_-_36421626 | 0.35 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr5_+_135496675 | 0.35 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr1_+_65730385 | 0.34 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr4_-_74864386 | 0.34 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chr7_+_73442102 | 0.34 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chr11_+_112832202 | 0.33 |
ENST00000534015.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr20_+_31823792 | 0.33 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr9_+_116263639 | 0.33 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr21_-_36421535 | 0.33 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr6_-_49712123 | 0.33 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr2_+_181845843 | 0.32 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr7_-_14029283 | 0.32 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 3.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.6 | 1.9 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.4 | 1.9 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 2.0 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 4.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 1.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 2.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 1.3 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.2 | 0.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 5.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 5.1 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 1.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 11.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 5.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.8 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.8 | 5.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 8.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 1.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.6 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 0.9 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.3 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.9 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 3.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 3.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.8 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.7 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 0.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 2.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.1 | 1.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 5.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 5.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 2.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 6.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 5.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 7.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 3.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.7 | 2.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.6 | 1.9 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.6 | 1.8 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.5 | 1.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.5 | 2.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.4 | 0.8 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.4 | 1.1 | GO:0060577 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.4 | 4.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.3 | GO:0044407 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.3 | 1.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.8 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 0.9 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.3 | 0.9 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.3 | 3.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 0.8 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 0.8 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 1.9 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 1.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.8 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 | 1.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 2.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 1.3 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 1.7 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.5 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.2 | 0.6 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.5 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 1.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.5 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.7 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 | 1.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 0.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.0 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 4.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 6.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 1.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.8 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 1.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 2.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 1.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 4.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.8 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 0.3 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 2.0 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 2.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 1.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.9 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.9 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.5 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.7 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.5 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 1.5 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 8.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 1.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 1.9 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.9 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 1.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.8 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0019050 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 8.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 3.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 9.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 3.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 8.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 9.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 6.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.1 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |