Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
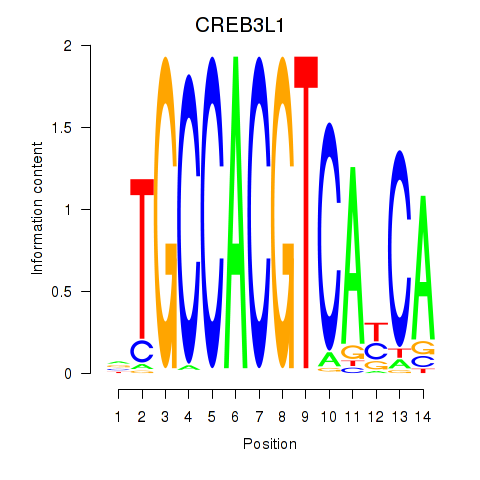
Results for CREB3L1_CREB3
Z-value: 0.82
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | CREB3L1 |
|
CREB3
|
ENSG00000107175.6 | CREB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L1 | hg19_v2_chr11_+_46299199_46299233 | 0.45 | 8.2e-02 | Click! |
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | 0.35 | 1.9e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_38864041 | 3.76 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr12_-_106641728 | 2.88 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr7_+_94023873 | 1.86 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr6_+_151561085 | 1.65 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr7_+_128379346 | 1.64 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr7_+_128379449 | 1.48 |
ENST00000479257.1 |
CALU |
calumenin |
| chr5_-_39425222 | 1.22 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_-_39425290 | 1.20 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_-_49395705 | 1.10 |
ENST00000419349.1 |
GPX1 |
glutathione peroxidase 1 |
| chr11_+_69455855 | 1.07 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr3_-_49395892 | 1.04 |
ENST00000419783.1 |
GPX1 |
glutathione peroxidase 1 |
| chr11_+_32112431 | 1.00 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr5_-_131562935 | 1.00 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr5_-_57756087 | 0.99 |
ENST00000274289.3 |
PLK2 |
polo-like kinase 2 |
| chr3_+_100211412 | 0.97 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chr5_-_39425068 | 0.93 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_111544254 | 0.90 |
ENST00000306732.3 |
PITX2 |
paired-like homeodomain 2 |
| chr5_-_131563501 | 0.84 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr5_-_93447333 | 0.76 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr7_-_30066233 | 0.73 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr3_+_50284321 | 0.72 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr16_+_56642489 | 0.71 |
ENST00000561491.1 |
MT2A |
metallothionein 2A |
| chr16_+_56642041 | 0.71 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr3_-_57583052 | 0.69 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr11_-_2906979 | 0.65 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr3_-_57583185 | 0.63 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr9_+_112542572 | 0.62 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chrX_-_10544942 | 0.61 |
ENST00000380779.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr3_-_57583130 | 0.61 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_-_121468513 | 0.60 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr17_-_10560619 | 0.59 |
ENST00000583535.1 |
MYH3 |
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr12_+_10365404 | 0.55 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr8_-_57906362 | 0.55 |
ENST00000262644.4 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
| chr12_-_56123444 | 0.53 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr19_+_49458107 | 0.52 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr3_-_121468602 | 0.51 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr1_+_160313062 | 0.50 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr1_+_160313165 | 0.49 |
ENST00000421914.1 ENST00000535857.1 ENST00000438008.1 |
NCSTN |
nicastrin |
| chr16_-_66959429 | 0.48 |
ENST00000420652.1 ENST00000299759.6 |
RRAD |
Ras-related associated with diabetes |
| chr17_-_7145475 | 0.47 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr3_-_156272924 | 0.46 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr12_+_72233487 | 0.45 |
ENST00000482439.2 ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15 |
TBC1 domain family, member 15 |
| chr12_-_49318715 | 0.44 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr17_+_57642886 | 0.44 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr3_+_133292574 | 0.44 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr13_+_113951532 | 0.42 |
ENST00000332556.4 |
LAMP1 |
lysosomal-associated membrane protein 1 |
| chr5_+_149340282 | 0.42 |
ENST00000286298.4 |
SLC26A2 |
solute carrier family 26 (anion exchanger), member 2 |
| chr20_-_17662878 | 0.42 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr9_+_114393634 | 0.42 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr1_-_101360331 | 0.41 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr15_-_40213080 | 0.41 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr1_+_145576007 | 0.41 |
ENST00000369298.1 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr1_+_145575980 | 0.40 |
ENST00000393045.2 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr13_-_36920615 | 0.40 |
ENST00000494062.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr13_-_36920872 | 0.39 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr8_+_64081214 | 0.39 |
ENST00000542911.2 |
YTHDF3 |
YTH domain family, member 3 |
| chr10_+_121410882 | 0.38 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr19_+_16187816 | 0.37 |
ENST00000588410.1 |
TPM4 |
tropomyosin 4 |
| chr6_+_31916733 | 0.37 |
ENST00000483004.1 |
CFB |
complement factor B |
| chrX_+_47441712 | 0.36 |
ENST00000218388.4 ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr17_+_40688190 | 0.35 |
ENST00000225927.2 |
NAGLU |
N-acetylglucosaminidase, alpha |
| chr4_-_119757239 | 0.33 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chrX_-_102941596 | 0.33 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr3_-_107941230 | 0.33 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr5_-_140700322 | 0.32 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr5_+_109025067 | 0.30 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chrX_+_153060090 | 0.30 |
ENST00000370086.3 ENST00000370085.3 |
SSR4 |
signal sequence receptor, delta |
| chr7_+_150929550 | 0.30 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr11_+_1891380 | 0.30 |
ENST00000429923.1 ENST00000418975.1 ENST00000406638.2 |
LSP1 |
lymphocyte-specific protein 1 |
| chr8_+_64081156 | 0.30 |
ENST00000517371.1 |
YTHDF3 |
YTH domain family, member 3 |
| chr13_-_36920420 | 0.30 |
ENST00000438666.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr8_+_22423168 | 0.29 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr20_-_17662705 | 0.29 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr19_-_48894104 | 0.29 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr11_-_67276100 | 0.29 |
ENST00000301488.3 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr19_-_15236562 | 0.28 |
ENST00000263383.3 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr4_+_56262115 | 0.28 |
ENST00000506198.1 ENST00000381334.5 ENST00000542052.1 |
TMEM165 |
transmembrane protein 165 |
| chr5_-_172198190 | 0.28 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr15_-_72668185 | 0.28 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr3_+_35721106 | 0.28 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_54121261 | 0.27 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr5_-_114961858 | 0.26 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr13_+_113951607 | 0.26 |
ENST00000397181.3 |
LAMP1 |
lysosomal-associated membrane protein 1 |
| chr12_-_50616382 | 0.26 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr7_+_39663061 | 0.25 |
ENST00000005257.2 |
RALA |
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr10_-_30348439 | 0.24 |
ENST00000375377.1 |
KIAA1462 |
KIAA1462 |
| chr7_-_100860851 | 0.24 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr9_-_98079965 | 0.24 |
ENST00000289081.3 |
FANCC |
Fanconi anemia, complementation group C |
| chr12_-_50616122 | 0.23 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr11_-_65686586 | 0.23 |
ENST00000438576.2 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chrX_+_55744228 | 0.23 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chrX_+_55744166 | 0.22 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr8_+_64081118 | 0.22 |
ENST00000539294.1 |
YTHDF3 |
YTH domain family, member 3 |
| chr1_-_160313025 | 0.22 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr1_-_241520525 | 0.22 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr3_+_122785895 | 0.22 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr4_-_54930790 | 0.21 |
ENST00000263921.3 |
CHIC2 |
cysteine-rich hydrophobic domain 2 |
| chr17_-_80231557 | 0.21 |
ENST00000392334.2 ENST00000314028.6 |
CSNK1D |
casein kinase 1, delta |
| chr11_-_62341445 | 0.21 |
ENST00000329251.4 |
EEF1G |
eukaryotic translation elongation factor 1 gamma |
| chr2_+_183580954 | 0.20 |
ENST00000264065.7 |
DNAJC10 |
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr1_+_11866207 | 0.20 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr1_-_65432171 | 0.19 |
ENST00000342505.4 |
JAK1 |
Janus kinase 1 |
| chrX_+_153059608 | 0.19 |
ENST00000370087.1 |
SSR4 |
signal sequence receptor, delta |
| chr10_-_111683308 | 0.19 |
ENST00000502935.1 ENST00000322238.8 ENST00000369680.4 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr19_-_15236470 | 0.18 |
ENST00000533747.1 ENST00000598709.1 ENST00000534378.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr22_+_39916558 | 0.18 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr12_+_56110247 | 0.18 |
ENST00000551926.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr9_-_127703333 | 0.18 |
ENST00000373555.4 |
GOLGA1 |
golgin A1 |
| chr3_+_37903432 | 0.18 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr14_+_50087468 | 0.17 |
ENST00000305386.2 |
MGAT2 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr15_+_44084040 | 0.17 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr5_-_10761206 | 0.17 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr19_+_50353944 | 0.17 |
ENST00000594151.1 ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr5_-_94890648 | 0.17 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr10_-_98346801 | 0.16 |
ENST00000371142.4 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
| chr8_+_133787586 | 0.16 |
ENST00000395379.1 ENST00000395386.2 ENST00000337920.4 |
PHF20L1 |
PHD finger protein 20-like 1 |
| chr19_-_54619006 | 0.16 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr12_-_125399573 | 0.15 |
ENST00000339647.5 |
UBC |
ubiquitin C |
| chr8_-_71519889 | 0.15 |
ENST00000521425.1 |
TRAM1 |
translocation associated membrane protein 1 |
| chr9_-_74980113 | 0.15 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr2_-_98280383 | 0.15 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr7_-_129592700 | 0.15 |
ENST00000472396.1 ENST00000355621.3 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr2_-_33824382 | 0.15 |
ENST00000238823.8 |
FAM98A |
family with sequence similarity 98, member A |
| chr19_-_15236173 | 0.15 |
ENST00000527093.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr8_+_140943416 | 0.15 |
ENST00000507535.3 |
C8orf17 |
chromosome 8 open reading frame 17 |
| chr11_-_65686496 | 0.15 |
ENST00000449692.3 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr2_+_28615669 | 0.15 |
ENST00000379619.1 ENST00000264716.4 |
FOSL2 |
FOS-like antigen 2 |
| chr19_+_50354393 | 0.14 |
ENST00000391842.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr19_-_54618650 | 0.14 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr17_-_66951474 | 0.14 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr19_-_4670345 | 0.14 |
ENST00000599630.1 ENST00000262947.3 |
C19orf10 |
chromosome 19 open reading frame 10 |
| chr17_-_19771216 | 0.13 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chrX_-_153059958 | 0.13 |
ENST00000370092.3 ENST00000217901.5 |
IDH3G |
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr10_-_111683183 | 0.13 |
ENST00000403138.2 ENST00000369683.1 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr2_-_33824336 | 0.13 |
ENST00000431950.1 ENST00000403368.1 ENST00000441530.2 |
FAM98A |
family with sequence similarity 98, member A |
| chr3_-_139108475 | 0.12 |
ENST00000515006.1 ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2 |
coatomer protein complex, subunit beta 2 (beta prime) |
| chr11_+_65686802 | 0.12 |
ENST00000376991.2 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_+_118443098 | 0.12 |
ENST00000392859.3 ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1 |
archain 1 |
| chr11_-_67407031 | 0.12 |
ENST00000335385.3 |
TBX10 |
T-box 10 |
| chr5_-_54603368 | 0.12 |
ENST00000508346.1 ENST00000251636.5 |
DHX29 |
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr15_+_44084503 | 0.11 |
ENST00000409960.2 ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2 |
small EDRK-rich factor 2 |
| chr3_-_28390581 | 0.11 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr11_-_119234876 | 0.11 |
ENST00000525735.1 |
USP2 |
ubiquitin specific peptidase 2 |
| chrX_-_153236819 | 0.11 |
ENST00000354233.3 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chr8_+_142138711 | 0.11 |
ENST00000518347.1 ENST00000262585.2 ENST00000424248.1 ENST00000519811.1 ENST00000520986.1 ENST00000523058.1 |
DENND3 |
DENN/MADD domain containing 3 |
| chr19_-_52097613 | 0.11 |
ENST00000301439.3 |
AC018755.1 |
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr10_-_27444143 | 0.11 |
ENST00000477432.1 |
YME1L1 |
YME1-like 1 ATPase |
| chr9_-_131709858 | 0.11 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr12_+_56552128 | 0.11 |
ENST00000548580.1 ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr19_+_2841433 | 0.11 |
ENST00000334241.4 ENST00000585966.1 ENST00000591539.1 |
ZNF555 |
zinc finger protein 555 |
| chr15_+_84904525 | 0.10 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr20_+_57267669 | 0.10 |
ENST00000356091.6 |
NPEPL1 |
aminopeptidase-like 1 |
| chr19_-_18391708 | 0.10 |
ENST00000600972.1 |
JUND |
jun D proto-oncogene |
| chr2_+_220042933 | 0.10 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chr8_+_22422749 | 0.10 |
ENST00000523900.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr1_+_44445549 | 0.10 |
ENST00000356836.6 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr16_-_86588627 | 0.09 |
ENST00000565482.1 ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD |
methenyltetrahydrofolate synthetase domain containing |
| chr7_+_139025875 | 0.09 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chrX_+_101854426 | 0.09 |
ENST00000536530.1 ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
| chr17_+_4046462 | 0.09 |
ENST00000577075.2 ENST00000575251.1 ENST00000301391.3 |
CYB5D2 |
cytochrome b5 domain containing 2 |
| chr5_+_126853301 | 0.09 |
ENST00000296666.8 ENST00000442138.2 ENST00000512635.2 |
PRRC1 |
proline-rich coiled-coil 1 |
| chr1_+_110162448 | 0.09 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr12_-_120638902 | 0.09 |
ENST00000551150.1 ENST00000313104.5 ENST00000547191.1 ENST00000546989.1 ENST00000392514.4 ENST00000228306.4 ENST00000550856.1 |
RPLP0 |
ribosomal protein, large, P0 |
| chr1_-_43833628 | 0.09 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr13_+_49822041 | 0.09 |
ENST00000538056.1 ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
| chr19_+_41256764 | 0.09 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr7_-_93633658 | 0.09 |
ENST00000433727.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr3_-_134204815 | 0.09 |
ENST00000514612.1 ENST00000510994.1 ENST00000354910.5 |
ANAPC13 |
anaphase promoting complex subunit 13 |
| chr3_-_119182523 | 0.09 |
ENST00000319172.5 |
TMEM39A |
transmembrane protein 39A |
| chr1_+_11866270 | 0.09 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr3_+_134205000 | 0.09 |
ENST00000512894.1 ENST00000513612.2 ENST00000606977.1 |
CEP63 |
centrosomal protein 63kDa |
| chrX_+_153237740 | 0.09 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr17_-_7155274 | 0.09 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr11_+_65686728 | 0.08 |
ENST00000312515.2 ENST00000525501.1 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr3_+_134204881 | 0.08 |
ENST00000511574.1 ENST00000337090.3 ENST00000383229.3 |
CEP63 |
centrosomal protein 63kDa |
| chr11_+_809961 | 0.08 |
ENST00000530797.1 |
RPLP2 |
ribosomal protein, large, P2 |
| chr1_+_228327923 | 0.08 |
ENST00000391865.3 |
GUK1 |
guanylate kinase 1 |
| chr7_+_140396946 | 0.08 |
ENST00000476470.1 ENST00000471136.1 |
NDUFB2 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr17_+_73089382 | 0.08 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr13_+_27998681 | 0.08 |
ENST00000381140.4 |
GTF3A |
general transcription factor IIIA |
| chr1_+_32645269 | 0.08 |
ENST00000373610.3 |
TXLNA |
taxilin alpha |
| chr6_+_127588020 | 0.08 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr19_+_18668572 | 0.08 |
ENST00000540691.1 ENST00000539106.1 ENST00000222307.4 |
KXD1 |
KxDL motif containing 1 |
| chr11_-_14521379 | 0.08 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr19_+_49122548 | 0.07 |
ENST00000245222.4 ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2 |
sphingosine kinase 2 |
| chrX_+_101854096 | 0.07 |
ENST00000246174.2 ENST00000537008.1 ENST00000541409.1 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
| chr6_+_116692102 | 0.07 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr5_-_131132614 | 0.07 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr6_-_135375921 | 0.07 |
ENST00000367820.2 ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L |
HBS1-like (S. cerevisiae) |
| chr15_-_72668805 | 0.07 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr12_-_125398850 | 0.07 |
ENST00000535859.1 ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC |
ubiquitin C |
| chr2_+_27255806 | 0.07 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr7_-_93633684 | 0.06 |
ENST00000222547.3 ENST00000425626.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr11_-_65624415 | 0.06 |
ENST00000524553.1 ENST00000527344.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr22_-_20850128 | 0.06 |
ENST00000328879.4 |
KLHL22 |
kelch-like family member 22 |
| chr10_-_97416400 | 0.06 |
ENST00000371224.2 ENST00000371221.3 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
| chr7_+_100271355 | 0.06 |
ENST00000436220.1 ENST00000424361.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_+_92414928 | 0.06 |
ENST00000362005.3 ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT |
bromodomain, testis-specific |
| chr5_-_32174369 | 0.06 |
ENST00000265070.6 |
GOLPH3 |
golgi phosphoprotein 3 (coat-protein) |
| chr19_+_51358166 | 0.05 |
ENST00000601503.1 ENST00000326003.2 ENST00000597286.1 ENST00000597483.1 |
KLK3 |
kallikrein-related peptidase 3 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 1.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.5 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 1.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.4 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 0.3 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 2.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.4 | 2.1 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 | 4.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 0.9 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.3 | 1.4 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 1.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 1.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 0.5 | GO:1902445 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.2 | 0.7 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 1.9 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.6 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 1.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.4 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.8 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.1 | 0.3 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.1 | 0.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.3 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.5 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 5.9 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 3.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.0 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 4.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 4.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 3.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.2 | 3.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 1.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0019012 | virion(GO:0019012) virion part(GO:0044423) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |