Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
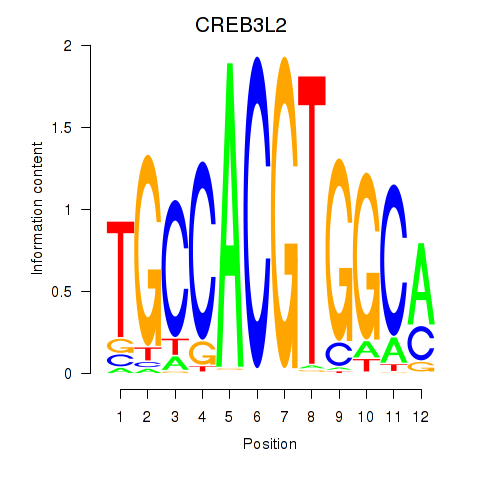
Results for CREB3L2
Z-value: 1.04
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | CREB3L2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg19_v2_chr7_-_137686791_137686821 | 0.84 | 4.4e-05 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_61584026 | 4.06 |
ENST00000370351.4 ENST00000370349.3 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr6_+_151561506 | 2.88 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_+_151561085 | 2.56 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr2_+_46926326 | 2.03 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr12_-_49318715 | 1.92 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr13_+_98794810 | 1.74 |
ENST00000595437.1 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr3_-_123603137 | 1.73 |
ENST00000360304.3 ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK |
myosin light chain kinase |
| chr5_-_150537279 | 1.45 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr16_-_88923285 | 1.44 |
ENST00000542788.1 ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr6_-_32811771 | 1.41 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr17_-_39968855 | 1.35 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr15_+_58724184 | 1.32 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr10_+_89419370 | 1.30 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr17_+_40688190 | 1.27 |
ENST00000225927.2 |
NAGLU |
N-acetylglucosaminidase, alpha |
| chr8_-_134584092 | 1.24 |
ENST00000522652.1 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chrX_+_105937068 | 1.22 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr17_-_26695013 | 1.22 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr1_-_27240455 | 1.18 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr8_-_134584152 | 1.18 |
ENST00000521180.1 ENST00000517668.1 ENST00000319914.5 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_+_32112431 | 1.17 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr17_-_26694979 | 1.16 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr2_+_46926048 | 1.12 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr17_+_39969183 | 1.11 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr7_-_134143841 | 1.05 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr10_+_131265443 | 1.03 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr1_+_11866207 | 1.03 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr1_+_180123969 | 1.00 |
ENST00000367602.3 ENST00000367600.5 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr6_+_32811885 | 0.99 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_-_74551172 | 0.98 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr22_+_38864041 | 0.97 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr19_+_11350278 | 0.96 |
ENST00000252453.8 |
C19orf80 |
chromosome 19 open reading frame 80 |
| chr19_+_49458107 | 0.93 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr9_+_706842 | 0.92 |
ENST00000382293.3 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr13_+_35516390 | 0.88 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr3_+_122785895 | 0.86 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr14_-_74551096 | 0.83 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr4_-_83812248 | 0.82 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr4_-_83812402 | 0.82 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr22_+_39916558 | 0.81 |
ENST00000337304.2 ENST00000396680.1 |
ATF4 |
activating transcription factor 4 |
| chr8_+_22224760 | 0.80 |
ENST00000359741.5 ENST00000520644.1 ENST00000240095.6 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr15_-_72668185 | 0.79 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr20_-_17662878 | 0.78 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr17_+_19552036 | 0.77 |
ENST00000581518.1 ENST00000395575.2 ENST00000584332.2 ENST00000339618.4 ENST00000579855.1 |
ALDH3A2 |
aldehyde dehydrogenase 3 family, member A2 |
| chr7_-_6523688 | 0.75 |
ENST00000490996.1 |
KDELR2 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr6_-_7911042 | 0.73 |
ENST00000379757.4 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr16_+_66914264 | 0.67 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr20_+_44519948 | 0.66 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chr17_+_78075361 | 0.66 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr22_-_43253189 | 0.64 |
ENST00000437119.2 ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
| chr1_+_110162448 | 0.63 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr20_+_44520009 | 0.61 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr3_-_57583052 | 0.61 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr1_+_11866270 | 0.58 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr1_-_45956822 | 0.57 |
ENST00000372086.3 ENST00000341771.6 |
TESK2 |
testis-specific kinase 2 |
| chr10_-_126849588 | 0.57 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr3_+_148709128 | 0.55 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr5_+_133984462 | 0.55 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr1_-_241520525 | 0.54 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr10_-_97416400 | 0.54 |
ENST00000371224.2 ENST00000371221.3 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
| chr7_-_6523755 | 0.53 |
ENST00000436575.1 ENST00000258739.4 |
DAGLB KDELR2 |
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr16_-_30134524 | 0.52 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr16_-_30134266 | 0.52 |
ENST00000484663.1 ENST00000478356.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr20_-_4795747 | 0.51 |
ENST00000379376.2 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr1_-_154193009 | 0.50 |
ENST00000368518.1 ENST00000368519.1 ENST00000368521.5 |
C1orf43 |
chromosome 1 open reading frame 43 |
| chr3_-_57583130 | 0.50 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr19_-_41256207 | 0.48 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr15_+_89181974 | 0.48 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr17_-_74722536 | 0.48 |
ENST00000585429.1 |
JMJD6 |
jumonji domain containing 6 |
| chr4_+_128651530 | 0.47 |
ENST00000281154.4 |
SLC25A31 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr2_-_73340146 | 0.46 |
ENST00000258098.6 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
| chr20_-_33413416 | 0.46 |
ENST00000359003.2 |
NCOA6 |
nuclear receptor coactivator 6 |
| chr11_+_67776012 | 0.46 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr15_-_72668805 | 0.45 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr2_+_207024306 | 0.44 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr11_-_57282349 | 0.43 |
ENST00000528450.1 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr17_+_72428218 | 0.43 |
ENST00000392628.2 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_6502580 | 0.43 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr17_-_74722672 | 0.42 |
ENST00000397625.4 ENST00000445478.2 |
JMJD6 |
jumonji domain containing 6 |
| chr8_+_56014949 | 0.41 |
ENST00000327381.6 |
XKR4 |
XK, Kell blood group complex subunit-related family, member 4 |
| chr2_-_69614373 | 0.41 |
ENST00000361060.5 ENST00000357308.4 |
GFPT1 |
glutamine--fructose-6-phosphate transaminase 1 |
| chr16_-_21314360 | 0.40 |
ENST00000219599.3 ENST00000576703.1 |
CRYM |
crystallin, mu |
| chr2_-_27341765 | 0.39 |
ENST00000405600.1 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
| chr2_+_241526126 | 0.37 |
ENST00000391984.2 ENST00000391982.2 ENST00000404753.3 ENST00000270364.7 ENST00000352879.4 ENST00000354082.4 |
CAPN10 |
calpain 10 |
| chr17_-_40333150 | 0.36 |
ENST00000264661.3 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr9_-_113800981 | 0.36 |
ENST00000538760.1 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr2_+_172543967 | 0.35 |
ENST00000534253.2 ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr3_+_148709310 | 0.35 |
ENST00000484197.1 ENST00000492285.2 ENST00000461191.1 |
GYG1 |
glycogenin 1 |
| chr19_-_47164386 | 0.35 |
ENST00000391916.2 ENST00000410105.2 |
DACT3 |
dishevelled-binding antagonist of beta-catenin 3 |
| chr1_+_231376941 | 0.35 |
ENST00000436239.1 ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT |
glyceronephosphate O-acyltransferase |
| chr2_-_101767715 | 0.34 |
ENST00000376840.4 ENST00000409318.1 |
TBC1D8 |
TBC1 domain family, member 8 (with GRAM domain) |
| chr2_+_172544011 | 0.33 |
ENST00000508530.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr17_-_7137857 | 0.32 |
ENST00000005340.5 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr7_+_150929550 | 0.32 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr2_-_47142884 | 0.32 |
ENST00000409105.1 ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr17_-_40333099 | 0.32 |
ENST00000607371.1 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr1_+_46016703 | 0.32 |
ENST00000481885.1 ENST00000351829.4 ENST00000471651.1 |
AKR1A1 |
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr14_+_105155925 | 0.32 |
ENST00000330634.7 ENST00000398337.4 ENST00000392634.4 |
INF2 |
inverted formin, FH2 and WH2 domain containing |
| chr5_+_34656569 | 0.31 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr20_+_2821340 | 0.31 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr1_+_42922173 | 0.31 |
ENST00000455780.1 ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr6_+_87865262 | 0.30 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr15_-_40213080 | 0.29 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr9_-_113800341 | 0.29 |
ENST00000358883.4 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr1_-_1167411 | 0.29 |
ENST00000263741.7 |
SDF4 |
stromal cell derived factor 4 |
| chr6_+_158733692 | 0.29 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr5_-_114880533 | 0.28 |
ENST00000274457.3 |
FEM1C |
fem-1 homolog c (C. elegans) |
| chr2_+_217498105 | 0.28 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr2_-_27341872 | 0.28 |
ENST00000312734.4 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
| chr17_-_7137582 | 0.27 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr7_+_101917407 | 0.27 |
ENST00000487284.1 |
CUX1 |
cut-like homeobox 1 |
| chr5_+_34656331 | 0.27 |
ENST00000265109.3 |
RAI14 |
retinoic acid induced 14 |
| chr22_-_43411106 | 0.27 |
ENST00000453643.1 ENST00000263246.3 ENST00000337959.4 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_+_172544182 | 0.26 |
ENST00000409197.1 ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr7_-_100171270 | 0.26 |
ENST00000538735.1 |
SAP25 |
Sin3A-associated protein, 25kDa |
| chr11_-_6502534 | 0.26 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr2_-_47143160 | 0.26 |
ENST00000409800.1 ENST00000409218.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr2_+_233562015 | 0.25 |
ENST00000427233.1 ENST00000373566.3 ENST00000373563.4 ENST00000428883.1 ENST00000456491.1 ENST00000409480.1 ENST00000421433.1 ENST00000425040.1 ENST00000430720.1 ENST00000409547.1 ENST00000423659.1 ENST00000409196.3 ENST00000409451.3 ENST00000429187.1 ENST00000440945.1 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
| chr1_+_42921761 | 0.25 |
ENST00000372562.1 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr14_+_101193164 | 0.24 |
ENST00000341267.4 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr12_-_48152428 | 0.24 |
ENST00000449771.2 ENST00000395358.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_+_22102611 | 0.24 |
ENST00000306433.4 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr6_+_167525277 | 0.24 |
ENST00000400926.2 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr2_+_172544294 | 0.23 |
ENST00000358002.6 ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_+_121416489 | 0.23 |
ENST00000541395.1 ENST00000544413.1 |
HNF1A |
HNF1 homeobox A |
| chr1_-_11866034 | 0.22 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr5_+_112196919 | 0.21 |
ENST00000505459.1 ENST00000282999.3 ENST00000515463.1 |
SRP19 |
signal recognition particle 19kDa |
| chr12_+_6833237 | 0.21 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr2_+_198365095 | 0.21 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr1_+_29138654 | 0.21 |
ENST00000234961.2 |
OPRD1 |
opioid receptor, delta 1 |
| chr19_+_34663397 | 0.21 |
ENST00000540746.2 ENST00000544216.3 ENST00000433627.5 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr1_-_11865982 | 0.21 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_-_27341966 | 0.20 |
ENST00000402394.1 ENST00000402550.1 ENST00000260595.5 |
CGREF1 |
cell growth regulator with EF-hand domain 1 |
| chrX_+_153686614 | 0.20 |
ENST00000369682.3 |
PLXNA3 |
plexin A3 |
| chr3_-_57583185 | 0.20 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr4_+_37455536 | 0.19 |
ENST00000381980.4 ENST00000508175.1 |
C4orf19 |
chromosome 4 open reading frame 19 |
| chr20_-_57607347 | 0.19 |
ENST00000395663.1 ENST00000395659.1 ENST00000243997.3 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr17_-_37308824 | 0.19 |
ENST00000415163.1 ENST00000441877.1 ENST00000444911.2 |
PLXDC1 |
plexin domain containing 1 |
| chr2_+_172543919 | 0.18 |
ENST00000452242.1 ENST00000340296.4 |
DYNC1I2 |
dynein, cytoplasmic 1, intermediate chain 2 |
| chr19_+_13875316 | 0.18 |
ENST00000319545.8 ENST00000593245.1 ENST00000040663.6 |
MRI1 |
methylthioribose-1-phosphate isomerase 1 |
| chr2_-_27545921 | 0.18 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chrX_+_55744228 | 0.17 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr19_+_1269324 | 0.17 |
ENST00000589710.1 ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP |
cold inducible RNA binding protein |
| chr15_-_90645679 | 0.17 |
ENST00000539790.1 ENST00000559482.1 ENST00000330062.3 |
IDH2 |
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr8_+_22102626 | 0.16 |
ENST00000519237.1 ENST00000397802.4 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr12_+_122064398 | 0.15 |
ENST00000330079.7 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr1_-_31712401 | 0.15 |
ENST00000373736.2 |
NKAIN1 |
Na+/K+ transporting ATPase interacting 1 |
| chr13_+_32889605 | 0.15 |
ENST00000380152.3 ENST00000544455.1 ENST00000530893.2 |
BRCA2 |
breast cancer 2, early onset |
| chr8_-_133117512 | 0.15 |
ENST00000414222.1 |
HHLA1 |
HERV-H LTR-associating 1 |
| chr12_+_121416340 | 0.15 |
ENST00000257555.6 ENST00000400024.2 |
HNF1A |
HNF1 homeobox A |
| chr12_-_48152611 | 0.14 |
ENST00000389212.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr12_+_57623869 | 0.14 |
ENST00000414700.3 ENST00000557703.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr22_-_29663954 | 0.13 |
ENST00000216085.7 |
RHBDD3 |
rhomboid domain containing 3 |
| chr5_-_143550159 | 0.13 |
ENST00000448443.2 ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5 |
Yip1 domain family, member 5 |
| chr11_-_62341445 | 0.12 |
ENST00000329251.4 |
EEF1G |
eukaryotic translation elongation factor 1 gamma |
| chr11_-_61560053 | 0.12 |
ENST00000537328.1 |
TMEM258 |
transmembrane protein 258 |
| chr12_+_6833437 | 0.12 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr10_+_103892787 | 0.12 |
ENST00000278070.2 ENST00000413464.2 |
PPRC1 |
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr1_+_29213678 | 0.12 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr1_+_92414952 | 0.11 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr15_+_59063478 | 0.11 |
ENST00000559228.1 ENST00000450403.2 |
FAM63B |
family with sequence similarity 63, member B |
| chr1_-_32801825 | 0.11 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr5_+_178368186 | 0.11 |
ENST00000320129.3 ENST00000519564.1 |
ZNF454 |
zinc finger protein 454 |
| chr6_-_36953833 | 0.10 |
ENST00000538808.1 ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1 |
mitochondrial carrier 1 |
| chr11_-_777467 | 0.10 |
ENST00000397472.2 ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1 |
Parkinson disease 7 domain containing 1 |
| chr1_+_44445549 | 0.10 |
ENST00000356836.6 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr8_-_144679296 | 0.10 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr6_-_84937314 | 0.09 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr20_-_44993012 | 0.09 |
ENST00000372229.1 ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr6_-_32140886 | 0.09 |
ENST00000395496.1 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr10_-_104474128 | 0.09 |
ENST00000260746.5 |
ARL3 |
ADP-ribosylation factor-like 3 |
| chr1_+_92414928 | 0.09 |
ENST00000362005.3 ENST00000370389.2 ENST00000399546.2 ENST00000423434.1 ENST00000394530.3 ENST00000440509.1 |
BRDT |
bromodomain, testis-specific |
| chr22_-_36903069 | 0.08 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr5_-_131132658 | 0.08 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr2_-_211036051 | 0.08 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr16_-_1525016 | 0.07 |
ENST00000262318.8 ENST00000448525.1 |
CLCN7 |
chloride channel, voltage-sensitive 7 |
| chrX_-_48433275 | 0.07 |
ENST00000376775.2 |
AC115618.1 |
Uncharacterized protein; cDNA FLJ26048 fis, clone PRS02384 |
| chr9_-_125667618 | 0.07 |
ENST00000423239.2 |
RC3H2 |
ring finger and CCCH-type domains 2 |
| chr12_+_57623477 | 0.07 |
ENST00000557487.1 ENST00000555634.1 ENST00000556689.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr20_-_18774614 | 0.07 |
ENST00000412553.1 |
LINC00652 |
long intergenic non-protein coding RNA 652 |
| chr17_+_72270429 | 0.07 |
ENST00000311014.6 |
DNAI2 |
dynein, axonemal, intermediate chain 2 |
| chr15_+_89182178 | 0.07 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr7_-_8302207 | 0.06 |
ENST00000407906.1 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr5_-_131132614 | 0.06 |
ENST00000307968.7 ENST00000307954.8 |
FNIP1 |
folliculin interacting protein 1 |
| chr10_+_43916052 | 0.05 |
ENST00000442526.2 |
RP11-517P14.2 |
RP11-517P14.2 |
| chr3_-_194119083 | 0.05 |
ENST00000401815.1 |
GP5 |
glycoprotein V (platelet) |
| chr1_+_32479430 | 0.05 |
ENST00000327300.7 ENST00000492989.1 |
KHDRBS1 |
KH domain containing, RNA binding, signal transduction associated 1 |
| chrX_+_55744166 | 0.05 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr2_+_74425689 | 0.05 |
ENST00000394053.2 ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr4_+_40058411 | 0.05 |
ENST00000261435.6 ENST00000515550.1 |
N4BP2 |
NEDD4 binding protein 2 |
| chr3_+_133292759 | 0.04 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chr16_+_333152 | 0.04 |
ENST00000219406.6 ENST00000404312.1 ENST00000456379.1 |
PDIA2 |
protein disulfide isomerase family A, member 2 |
| chr2_-_70520539 | 0.04 |
ENST00000482975.2 ENST00000438261.1 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
| chr7_-_100808843 | 0.04 |
ENST00000249330.2 |
VGF |
VGF nerve growth factor inducible |
| chr1_-_153940097 | 0.03 |
ENST00000413622.1 ENST00000310483.6 |
SLC39A1 |
solute carrier family 39 (zinc transporter), member 1 |
| chr12_-_76953573 | 0.03 |
ENST00000549646.1 ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr18_+_56530794 | 0.03 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr6_+_127587755 | 0.03 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr15_+_89182156 | 0.02 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr5_-_138725560 | 0.02 |
ENST00000412103.2 ENST00000457570.2 |
MZB1 |
marginal zone B and B1 cell-specific protein |
| chr12_+_57624119 | 0.02 |
ENST00000555773.1 ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 6.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 6.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.5 | 1.8 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.4 | 1.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.4 | 5.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.3 | 1.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 0.9 | GO:1902445 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.3 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 0.8 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 1.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.2 | 2.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.6 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 0.2 | 0.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 1.0 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 2.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 1.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.1 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.3 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 1.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.3 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 1.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.8 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.2 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.2 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 4.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 1.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.4 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 1.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 0.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.4 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.9 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.9 | GO:0008104 | protein localization(GO:0008104) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.5 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0016310 | phosphorylation(GO:0016310) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 5.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.3 | 0.8 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 1.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 1.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 0.9 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.2 | 0.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 0.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 2.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.9 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 1.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.2 | 1.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 3.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 1.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 1.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 2.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 2.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 3.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |