Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for CUACAGU
Z-value: 0.24
miRNA associated with seed CUACAGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-139-5p
|
MIMAT0000250 |
Activity profile of CUACAGU motif
Sorted Z-values of CUACAGU motif
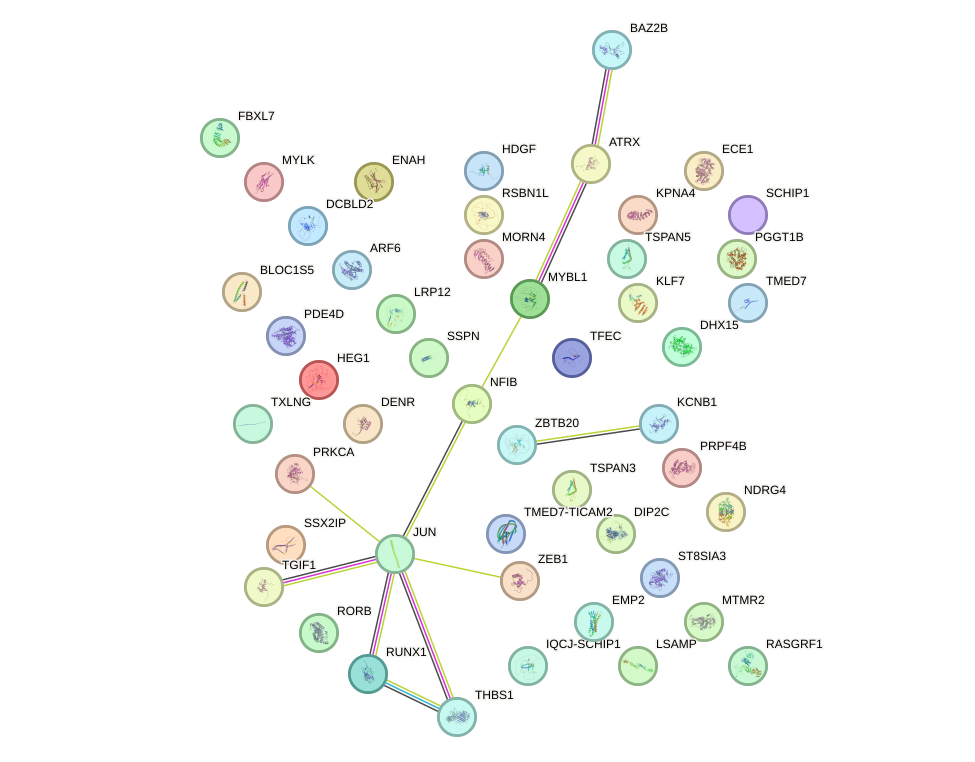
Network of associatons between targets according to the STRING database.
First level regulatory network of CUACAGU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_136834982 | 0.34 |
ENST00000510689.1 ENST00000394945.1 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr15_+_39873268 | 0.31 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr3_+_159557637 | 0.24 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr3_+_158991025 | 0.23 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr10_+_31608054 | 0.17 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr3_-_114790179 | 0.15 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_-_225840747 | 0.15 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr4_-_99579733 | 0.13 |
ENST00000305798.3 |
TSPAN5 |
tetraspanin 5 |
| chr9_-_14314066 | 0.12 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr5_-_127873659 | 0.12 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr16_+_58497567 | 0.11 |
ENST00000258187.5 |
NDRG4 |
NDRG family member 4 |
| chr8_-_67525473 | 0.11 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr3_-_123603137 | 0.11 |
ENST00000360304.3 ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK |
myosin light chain kinase |
| chr16_-_10674528 | 0.10 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr5_+_15500280 | 0.09 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr12_+_26348429 | 0.09 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr18_-_53255766 | 0.09 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr10_-_735553 | 0.07 |
ENST00000280886.6 ENST00000423550.1 |
DIP2C |
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr3_-_124774802 | 0.07 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr18_+_3451646 | 0.07 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr1_-_21671968 | 0.07 |
ENST00000415912.2 |
ECE1 |
endothelin converting enzyme 1 |
| chr15_-_77363513 | 0.06 |
ENST00000267970.4 |
TSPAN3 |
tetraspanin 3 |
| chr5_-_59189545 | 0.06 |
ENST00000340635.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr10_-_70287231 | 0.06 |
ENST00000609923.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr20_-_48099182 | 0.06 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr1_-_59249732 | 0.04 |
ENST00000371222.2 |
JUN |
jun proto-oncogene |
| chr3_-_98620500 | 0.04 |
ENST00000326840.6 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
| chr2_-_208030647 | 0.04 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr16_+_57662419 | 0.03 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_+_129939779 | 0.03 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr12_+_123237321 | 0.03 |
ENST00000280557.6 ENST00000455982.2 |
DENR |
density-regulated protein |
| chrX_-_77041685 | 0.02 |
ENST00000373344.5 ENST00000395603.3 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
| chr4_+_77870856 | 0.02 |
ENST00000264893.6 ENST00000502584.1 ENST00000510641.1 |
SEPT11 |
septin 11 |
| chr8_-_105601134 | 0.02 |
ENST00000276654.5 ENST00000424843.2 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
| chr9_+_77112244 | 0.02 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr6_+_4021554 | 0.02 |
ENST00000337659.6 |
PRPF4B |
pre-mRNA processing factor 4B |
| chr2_-_160472952 | 0.02 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chrX_+_16804544 | 0.02 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr3_-_160283348 | 0.02 |
ENST00000334256.4 |
KPNA4 |
karyopherin alpha 4 (importin alpha 3) |
| chr11_-_95657231 | 0.02 |
ENST00000409459.1 ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2 |
myotubularin related protein 2 |
| chr21_-_36260980 | 0.02 |
ENST00000344691.4 ENST00000358356.5 |
RUNX1 |
runt-related transcription factor 1 |
| chr2_+_181845298 | 0.01 |
ENST00000410062.4 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr7_-_115670792 | 0.01 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr10_+_82213904 | 0.01 |
ENST00000429989.3 |
TSPAN14 |
tetraspanin 14 |
| chr5_-_114598548 | 0.01 |
ENST00000379615.3 ENST00000419445.1 |
PGGT1B |
protein geranylgeranyltransferase type I, beta subunit |
| chr6_-_8064567 | 0.01 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr15_-_79383102 | 0.01 |
ENST00000558480.2 ENST00000419573.3 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr1_-_85156216 | 0.01 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_1822495 | 0.01 |
ENST00000378609.4 |
GNB1 |
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr10_-_99393242 | 0.01 |
ENST00000370635.3 ENST00000478953.1 ENST00000335628.3 |
MORN4 |
MORN repeat containing 4 |
| chr7_+_77325738 | 0.01 |
ENST00000334955.8 |
RSBN1L |
round spermatid basic protein 1-like |
| chr18_+_55018044 | 0.00 |
ENST00000324000.3 |
ST8SIA3 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_-_156721502 | 0.00 |
ENST00000357325.5 |
HDGF |
hepatoma-derived growth factor |
| chr12_+_123868320 | 0.00 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr14_+_50359773 | 0.00 |
ENST00000298316.5 |
ARF6 |
ADP-ribosylation factor 6 |
| chr3_-_116164306 | 0.00 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr5_-_114961858 | 0.00 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr4_-_24586140 | 0.00 |
ENST00000336812.4 |
DHX15 |
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr17_+_64298944 | 0.00 |
ENST00000413366.3 |
PRKCA |
protein kinase C, alpha |
| chr19_+_10527449 | 0.00 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr15_+_57210818 | 0.00 |
ENST00000438423.2 ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12 |
transcription factor 12 |
| chr1_-_149982624 | 0.00 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0090270 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |