Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for CUX2
Z-value: 1.15
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.9 | CUX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg19_v2_chr12_+_111471828_111471975 | 0.50 | 4.9e-02 | Click! |
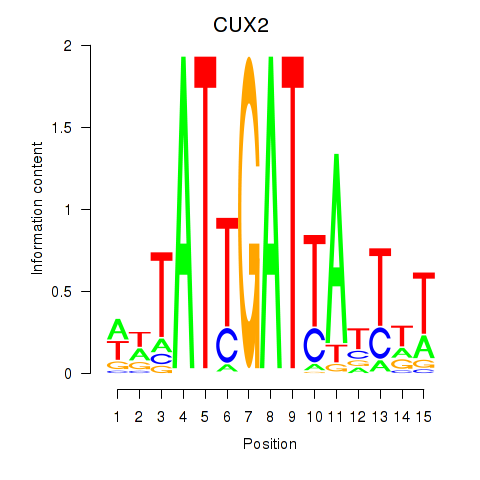
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_64216748 | 3.45 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_74275057 | 3.24 |
ENST00000511370.1 |
ALB |
albumin |
| chr4_+_69681710 | 3.23 |
ENST00000265403.7 ENST00000458688.2 |
UGT2B10 |
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr4_+_69962212 | 2.63 |
ENST00000508661.1 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr4_+_69962185 | 2.63 |
ENST00000305231.7 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_-_28634874 | 2.33 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_45409011 | 2.27 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr3_+_157827841 | 1.93 |
ENST00000295930.3 ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chr13_-_46679185 | 1.90 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr17_-_64225508 | 1.79 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr8_-_38008783 | 1.74 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr11_+_22689648 | 1.70 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr13_-_46679144 | 1.54 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr18_+_29171689 | 1.51 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr1_-_109935819 | 1.40 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr1_+_207262627 | 1.33 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr20_-_22566089 | 1.30 |
ENST00000377115.4 |
FOXA2 |
forkhead box A2 |
| chr1_+_207262578 | 1.28 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr20_+_34802295 | 1.25 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chrX_+_49178536 | 1.24 |
ENST00000442437.2 |
GAGE12J |
G antigen 12J |
| chr1_+_207262881 | 1.17 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr10_+_114135952 | 1.15 |
ENST00000356116.1 ENST00000433418.1 ENST00000354273.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chrX_+_49363665 | 1.13 |
ENST00000381700.6 |
GAGE1 |
G antigen 1 |
| chrX_+_49235708 | 1.08 |
ENST00000381725.1 |
GAGE2B |
G antigen 2B |
| chr3_-_120365866 | 1.07 |
ENST00000475447.2 |
HGD |
homogentisate 1,2-dioxygenase |
| chr2_+_211342432 | 1.06 |
ENST00000430249.2 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr4_-_70080449 | 1.06 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr6_+_26273144 | 1.01 |
ENST00000377733.2 |
HIST1H2BI |
histone cluster 1, H2bi |
| chr4_-_69817481 | 1.01 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_+_207262540 | 0.99 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr11_-_35440796 | 0.98 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr16_+_15489603 | 0.96 |
ENST00000568766.1 ENST00000287594.7 |
RP11-1021N1.1 MPV17L |
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr19_-_47287990 | 0.93 |
ENST00000593713.1 ENST00000598022.1 ENST00000434726.2 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr12_+_81471816 | 0.91 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chrX_-_11284095 | 0.91 |
ENST00000303025.6 ENST00000534860.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chrX_+_30261847 | 0.89 |
ENST00000378981.3 ENST00000397550.1 |
MAGEB1 |
melanoma antigen family B, 1 |
| chr7_+_120629653 | 0.89 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr19_-_36297632 | 0.86 |
ENST00000588266.2 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr19_+_42301079 | 0.82 |
ENST00000596544.1 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr8_+_77593448 | 0.82 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr3_-_194072019 | 0.80 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr4_-_144940477 | 0.77 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr4_+_155484155 | 0.75 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr11_+_57365150 | 0.74 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr13_-_46756351 | 0.71 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr7_-_5998714 | 0.70 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr3_+_157828152 | 0.69 |
ENST00000476899.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chrM_+_4431 | 0.68 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr11_-_35440579 | 0.68 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr8_+_77593474 | 0.65 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr1_+_198608146 | 0.64 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr9_+_35673853 | 0.64 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr12_+_19282643 | 0.63 |
ENST00000317589.4 ENST00000355397.3 ENST00000359180.3 ENST00000309364.4 ENST00000540972.1 ENST00000429027.2 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr8_+_94241867 | 0.63 |
ENST00000598428.1 |
AC016885.1 |
Uncharacterized protein |
| chr11_-_57158109 | 0.62 |
ENST00000525955.1 ENST00000533605.1 ENST00000311862.5 |
PRG2 |
proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) |
| chr4_-_145061788 | 0.61 |
ENST00000512064.1 ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA GYPB |
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr11_-_77734260 | 0.60 |
ENST00000353172.5 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
| chr8_-_86253888 | 0.59 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chrX_+_8432871 | 0.58 |
ENST00000381032.1 ENST00000453306.1 ENST00000444481.1 |
VCX3B |
variable charge, X-linked 3B |
| chr19_-_4535233 | 0.58 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr1_+_73771844 | 0.57 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr2_+_211421262 | 0.57 |
ENST00000233072.5 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr6_+_46761118 | 0.56 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr4_-_144826682 | 0.56 |
ENST00000358615.4 ENST00000437468.2 |
GYPE |
glycophorin E (MNS blood group) |
| chr16_-_18573396 | 0.56 |
ENST00000543392.1 ENST00000381474.3 ENST00000330537.6 |
NOMO2 |
NODAL modulator 2 |
| chr5_+_59726565 | 0.56 |
ENST00000412930.2 |
FKSG52 |
FKSG52 |
| chrX_+_9431324 | 0.54 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr6_+_30130969 | 0.54 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr21_-_37270727 | 0.53 |
ENST00000599809.1 |
FKSG68 |
FKSG68 |
| chr16_+_16326352 | 0.53 |
ENST00000399336.4 ENST00000263012.6 ENST00000538468.1 |
NOMO3 |
NODAL modulator 3 |
| chr9_-_110540419 | 0.53 |
ENST00000398726.3 |
AL162389.1 |
Uncharacterized protein |
| chr15_+_41057818 | 0.51 |
ENST00000558467.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr16_+_84801852 | 0.51 |
ENST00000569925.1 ENST00000567526.1 |
USP10 |
ubiquitin specific peptidase 10 |
| chr11_-_59633951 | 0.50 |
ENST00000257264.3 |
TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr16_+_33204156 | 0.50 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr6_-_26285737 | 0.50 |
ENST00000377727.1 ENST00000289352.1 |
HIST1H4H |
histone cluster 1, H4h |
| chr16_+_30064444 | 0.49 |
ENST00000395248.1 ENST00000566897.1 ENST00000568435.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr15_+_51973680 | 0.48 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr16_+_30064411 | 0.48 |
ENST00000338110.5 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr10_+_5238793 | 0.48 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr2_+_207220463 | 0.48 |
ENST00000598562.1 |
AC017081.1 |
Uncharacterized protein |
| chr9_-_138853156 | 0.47 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr8_-_108510224 | 0.47 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr3_+_25831567 | 0.47 |
ENST00000280701.3 ENST00000420173.2 |
OXSM |
3-oxoacyl-ACP synthase, mitochondrial |
| chr4_+_187187098 | 0.46 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr20_+_1099233 | 0.46 |
ENST00000246015.4 ENST00000335877.6 ENST00000438768.2 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr6_-_27114577 | 0.46 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr1_+_57320437 | 0.45 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr2_+_103035102 | 0.44 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr11_+_43380459 | 0.44 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr16_-_75467318 | 0.44 |
ENST00000283882.3 |
CFDP1 |
craniofacial development protein 1 |
| chr9_-_100881466 | 0.43 |
ENST00000341469.2 ENST00000342043.3 ENST00000375098.3 |
TRIM14 |
tripartite motif containing 14 |
| chr6_+_24667257 | 0.43 |
ENST00000537591.1 ENST00000230048.4 |
ACOT13 |
acyl-CoA thioesterase 13 |
| chr8_-_86290333 | 0.43 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr2_+_58655461 | 0.42 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr12_-_71182695 | 0.42 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr16_+_70258261 | 0.41 |
ENST00000594734.1 |
FKSG63 |
FKSG63 |
| chr5_-_133706695 | 0.41 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr15_-_58306295 | 0.41 |
ENST00000559517.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr8_-_91095099 | 0.40 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr11_+_101983176 | 0.40 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr5_-_115910630 | 0.39 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr19_+_36486078 | 0.39 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr12_+_6602517 | 0.39 |
ENST00000315579.5 ENST00000539714.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr15_+_58724184 | 0.38 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr6_+_56954808 | 0.38 |
ENST00000510483.1 ENST00000370706.4 ENST00000357489.3 |
ZNF451 |
zinc finger protein 451 |
| chr17_-_34890665 | 0.37 |
ENST00000586007.1 |
MYO19 |
myosin XIX |
| chrX_-_107975917 | 0.37 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr4_+_71263599 | 0.37 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr15_-_38519066 | 0.37 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr6_-_27799305 | 0.36 |
ENST00000357549.2 |
HIST1H4K |
histone cluster 1, H4k |
| chr17_+_34171081 | 0.36 |
ENST00000585577.1 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr7_+_16828866 | 0.36 |
ENST00000597084.1 |
AC073333.1 |
Uncharacterized protein |
| chr14_-_37642016 | 0.36 |
ENST00000331299.5 |
SLC25A21 |
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr3_+_39424828 | 0.36 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chrX_-_107682702 | 0.36 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr1_+_44870866 | 0.36 |
ENST00000355387.2 ENST00000361799.2 |
RNF220 |
ring finger protein 220 |
| chrX_-_101718085 | 0.35 |
ENST00000372750.1 ENST00000372752.1 |
NXF2B |
nuclear RNA export factor 2B |
| chr2_+_169312350 | 0.35 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr5_-_34916871 | 0.34 |
ENST00000382038.2 |
RAD1 |
RAD1 homolog (S. pombe) |
| chr5_+_54455946 | 0.34 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr11_-_57177586 | 0.34 |
ENST00000529411.1 |
RP11-872D17.8 |
Uncharacterized protein |
| chr11_+_101785727 | 0.33 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr14_+_21458127 | 0.33 |
ENST00000382985.4 ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17 |
methyltransferase like 17 |
| chr10_-_73975657 | 0.33 |
ENST00000394919.1 ENST00000526751.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chrX_+_101478829 | 0.32 |
ENST00000372763.1 ENST00000372758.1 |
NXF2 |
nuclear RNA export factor 2 |
| chr2_+_120687335 | 0.32 |
ENST00000544261.1 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr17_-_34890732 | 0.31 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr5_-_11588907 | 0.31 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr19_-_3786354 | 0.31 |
ENST00000395040.2 ENST00000310132.6 |
MATK |
megakaryocyte-associated tyrosine kinase |
| chr1_+_67632083 | 0.30 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chr11_-_104480019 | 0.30 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr2_-_27886460 | 0.30 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr21_-_43816152 | 0.30 |
ENST00000433957.2 ENST00000398397.3 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr19_-_13044494 | 0.30 |
ENST00000593021.1 ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_16083154 | 0.30 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr12_+_74931551 | 0.30 |
ENST00000519948.2 |
ATXN7L3B |
ataxin 7-like 3B |
| chr4_-_10686475 | 0.29 |
ENST00000226951.6 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr16_+_12059091 | 0.29 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr2_-_220264703 | 0.29 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr3_-_79068594 | 0.28 |
ENST00000436010.2 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr1_+_33439268 | 0.28 |
ENST00000594612.1 |
FKSG48 |
FKSG48 |
| chr16_-_2314222 | 0.28 |
ENST00000566397.1 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
| chr15_-_47426320 | 0.28 |
ENST00000557832.1 |
FKSG62 |
FKSG62 |
| chr16_-_70239683 | 0.28 |
ENST00000601706.1 |
AC009060.1 |
Uncharacterized protein |
| chr2_-_97405775 | 0.28 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr17_-_34890759 | 0.27 |
ENST00000431794.3 |
MYO19 |
myosin XIX |
| chr7_-_23510086 | 0.27 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr9_-_21187598 | 0.26 |
ENST00000421715.1 |
IFNA4 |
interferon, alpha 4 |
| chr2_+_27886330 | 0.26 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr17_-_72772462 | 0.26 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr3_+_52811596 | 0.25 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr8_-_141728760 | 0.25 |
ENST00000430260.2 |
PTK2 |
protein tyrosine kinase 2 |
| chr1_+_110546700 | 0.25 |
ENST00000359172.3 ENST00000393614.4 |
AHCYL1 |
adenosylhomocysteinase-like 1 |
| chr7_-_104909435 | 0.25 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr3_-_57233966 | 0.25 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr1_+_12976450 | 0.25 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr19_+_14672755 | 0.24 |
ENST00000594545.1 |
TECR |
trans-2,3-enoyl-CoA reductase |
| chr17_+_41132564 | 0.24 |
ENST00000361677.1 ENST00000589705.1 |
RUNDC1 |
RUN domain containing 1 |
| chr1_-_54304212 | 0.24 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr2_+_86333301 | 0.24 |
ENST00000254630.7 |
PTCD3 |
pentatricopeptide repeat domain 3 |
| chr2_+_102456277 | 0.24 |
ENST00000421882.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_+_50571949 | 0.24 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr13_+_48611665 | 0.24 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr9_-_21166659 | 0.23 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
| chr5_-_70316737 | 0.23 |
ENST00000194097.4 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr16_+_446713 | 0.23 |
ENST00000397722.1 ENST00000454619.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr22_+_24407642 | 0.23 |
ENST00000454754.1 ENST00000263119.5 |
CABIN1 |
calcineurin binding protein 1 |
| chr19_-_16008880 | 0.23 |
ENST00000011989.7 ENST00000221700.6 |
CYP4F2 |
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr6_-_11779014 | 0.23 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr22_+_24198890 | 0.23 |
ENST00000345044.6 |
SLC2A11 |
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr2_+_27255806 | 0.22 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr2_+_170655322 | 0.22 |
ENST00000260956.4 ENST00000417292.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr12_-_81763184 | 0.22 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr6_-_167571817 | 0.22 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr12_+_28410128 | 0.21 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr1_-_115259337 | 0.21 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr15_+_42651691 | 0.21 |
ENST00000357568.3 ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3 |
calpain 3, (p94) |
| chr16_-_28374829 | 0.20 |
ENST00000532254.1 |
NPIPB6 |
nuclear pore complex interacting protein family, member B6 |
| chr1_-_32384693 | 0.20 |
ENST00000602683.1 ENST00000470404.1 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr6_+_106534192 | 0.20 |
ENST00000369091.2 ENST00000369096.4 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr8_-_19459993 | 0.20 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr12_-_10978957 | 0.20 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr10_+_70980051 | 0.20 |
ENST00000354624.5 ENST00000395086.2 |
HKDC1 |
hexokinase domain containing 1 |
| chr14_+_97263641 | 0.20 |
ENST00000216639.3 |
VRK1 |
vaccinia related kinase 1 |
| chr17_+_7461580 | 0.20 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_-_11779174 | 0.20 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr21_-_43816052 | 0.19 |
ENST00000398405.1 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr5_-_180665195 | 0.19 |
ENST00000509148.1 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr3_+_67705121 | 0.19 |
ENST00000464420.1 ENST00000482677.1 |
RP11-81N13.1 |
RP11-81N13.1 |
| chr7_+_93535866 | 0.19 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr1_-_190446759 | 0.19 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr11_-_89653576 | 0.19 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr8_+_24151620 | 0.18 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr19_+_54466179 | 0.18 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr3_+_181429704 | 0.18 |
ENST00000431565.2 ENST00000325404.1 |
SOX2 |
SRY (sex determining region Y)-box 2 |
| chr6_-_11779403 | 0.18 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_+_155179012 | 0.18 |
ENST00000609421.1 |
MTX1 |
metaxin 1 |
| chr12_-_81763127 | 0.18 |
ENST00000541017.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_+_18230685 | 0.18 |
ENST00000340135.3 ENST00000534640.1 |
RP11-113D6.10 |
Putative mitochondrial carrier protein LOC494141 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.9 | 3.4 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.8 | 2.3 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.6 | 5.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.6 | 1.7 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.5 | 4.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.4 | 1.3 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.6 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 1.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.2 | 5.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.2 | 0.9 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 1.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 0.5 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.5 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 | 0.5 | GO:0071790 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 0.6 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 0.5 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.6 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 1.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 2.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 0.9 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.6 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.5 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.4 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.4 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.5 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.7 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0098904 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.9 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0070245 | snRNA transcription from RNA polymerase III promoter(GO:0042796) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 2.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0002834 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.4 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.3 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 2.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.5 | 1.6 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.3 | 2.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.3 | 9.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.9 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 3.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 0.5 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.2 | 1.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 4.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 1.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) NADH pyrophosphatase activity(GO:0035529) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.7 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 1.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.3 | 5.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.5 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 0.3 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 4.5 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |