Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
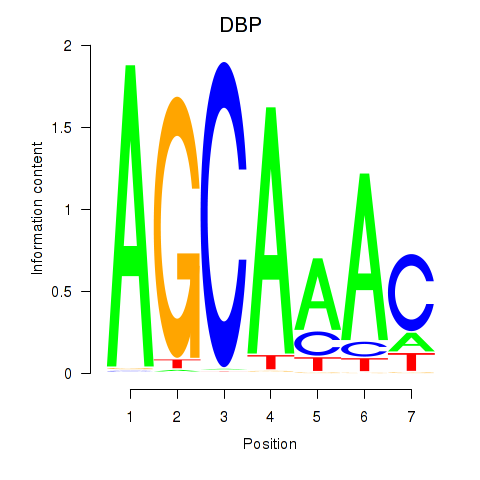
Results for DBP
Z-value: 0.94
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | DBP |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140692_49140709 | 0.43 | 9.9e-02 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_216257849 | 4.29 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr2_-_21266935 | 2.95 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr12_-_89746173 | 2.76 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr4_-_23891693 | 2.71 |
ENST00000264867.2 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr2_-_88427568 | 2.61 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr11_+_121461097 | 2.27 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr3_-_145878954 | 2.18 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr17_-_56492989 | 2.04 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chrX_+_23801280 | 2.00 |
ENST00000379251.3 ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
| chr22_+_38201114 | 1.83 |
ENST00000340857.2 |
H1F0 |
H1 histone family, member 0 |
| chr15_+_58702742 | 1.77 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr17_+_72426891 | 1.72 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr20_-_14318248 | 1.70 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr8_-_17533838 | 1.62 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr3_+_190333097 | 1.60 |
ENST00000412080.1 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chr3_-_52479043 | 1.57 |
ENST00000231721.2 ENST00000475739.1 |
SEMA3G |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr9_-_112083229 | 1.39 |
ENST00000374566.3 ENST00000374557.4 |
EPB41L4B |
erythrocyte membrane protein band 4.1 like 4B |
| chr3_-_185538849 | 1.30 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_-_22234381 | 1.27 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr20_+_11898507 | 1.24 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr8_+_104310661 | 1.15 |
ENST00000522566.1 |
FZD6 |
frizzled family receptor 6 |
| chr8_-_80993010 | 1.10 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr17_+_48624450 | 1.10 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr17_+_79953310 | 1.02 |
ENST00000582355.2 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr1_+_82266053 | 1.00 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr16_-_18468926 | 0.95 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_21593972 | 0.94 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr3_-_149375783 | 0.93 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr1_+_167298281 | 0.89 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr17_-_34308524 | 0.83 |
ENST00000293275.3 |
CCL16 |
chemokine (C-C motif) ligand 16 |
| chr5_-_16916624 | 0.82 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr5_-_111091948 | 0.82 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr20_+_361261 | 0.81 |
ENST00000217233.3 |
TRIB3 |
tribbles pseudokinase 3 |
| chr4_-_186732048 | 0.79 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr3_+_123813543 | 0.78 |
ENST00000360013.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr6_+_10528560 | 0.76 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr7_+_26331541 | 0.74 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr17_+_42429493 | 0.73 |
ENST00000586242.1 |
GRN |
granulin |
| chr13_+_97874574 | 0.67 |
ENST00000343600.4 ENST00000345429.6 ENST00000376673.3 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr2_-_208031943 | 0.65 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_129083772 | 0.64 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr1_+_28764653 | 0.63 |
ENST00000373836.3 |
PHACTR4 |
phosphatase and actin regulator 4 |
| chr13_+_97928395 | 0.63 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr2_+_234602305 | 0.61 |
ENST00000406651.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr14_+_58765103 | 0.60 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr7_-_121784285 | 0.59 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr17_+_2240775 | 0.58 |
ENST00000268989.3 ENST00000426855.2 |
SGSM2 |
small G protein signaling modulator 2 |
| chr15_+_90544532 | 0.57 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chr17_-_42580738 | 0.56 |
ENST00000585614.1 ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8 |
G patch domain containing 8 |
| chrX_+_107288197 | 0.54 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr1_+_155051305 | 0.51 |
ENST00000368408.3 |
EFNA3 |
ephrin-A3 |
| chr1_+_229440129 | 0.51 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chrX_+_107288239 | 0.50 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr7_+_99613212 | 0.48 |
ENST00000426572.1 ENST00000535170.1 |
ZKSCAN1 |
zinc finger with KRAB and SCAN domains 1 |
| chr1_-_92952433 | 0.48 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr4_+_185395947 | 0.47 |
ENST00000605834.1 |
RP11-326I11.3 |
RP11-326I11.3 |
| chr14_+_24590560 | 0.47 |
ENST00000558325.1 |
RP11-468E2.6 |
RP11-468E2.6 |
| chr5_+_180682720 | 0.46 |
ENST00000599439.1 |
AC008443.1 |
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr16_+_30960375 | 0.46 |
ENST00000318663.4 ENST00000566237.1 ENST00000562699.1 |
ORAI3 |
ORAI calcium release-activated calcium modulator 3 |
| chr17_+_28705921 | 0.45 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr22_-_31742218 | 0.45 |
ENST00000266269.5 ENST00000405309.3 ENST00000351933.4 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chrX_-_54070607 | 0.44 |
ENST00000338154.6 ENST00000338946.6 |
PHF8 |
PHD finger protein 8 |
| chr7_-_50633078 | 0.43 |
ENST00000444124.2 |
DDC |
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr10_-_29811456 | 0.43 |
ENST00000535393.1 |
SVIL |
supervillin |
| chr4_-_89978299 | 0.40 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr8_-_29120580 | 0.39 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chrX_-_10851762 | 0.39 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr17_+_38498594 | 0.39 |
ENST00000394081.3 |
RARA |
retinoic acid receptor, alpha |
| chr2_-_174830430 | 0.38 |
ENST00000310015.6 ENST00000455789.2 |
SP3 |
Sp3 transcription factor |
| chr7_-_129592471 | 0.37 |
ENST00000473814.2 ENST00000490974.1 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
| chr14_-_23479331 | 0.37 |
ENST00000397377.1 ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93 |
chromosome 14 open reading frame 93 |
| chr17_+_4613776 | 0.36 |
ENST00000269260.2 |
ARRB2 |
arrestin, beta 2 |
| chr18_-_51750948 | 0.35 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr10_-_100995540 | 0.34 |
ENST00000370546.1 ENST00000404542.1 |
HPSE2 |
heparanase 2 |
| chr4_-_185395672 | 0.33 |
ENST00000393593.3 |
IRF2 |
interferon regulatory factor 2 |
| chr18_-_25616519 | 0.33 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr10_-_100995603 | 0.33 |
ENST00000370552.3 ENST00000370549.1 |
HPSE2 |
heparanase 2 |
| chr2_-_152830479 | 0.33 |
ENST00000360283.6 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr4_-_157892498 | 0.32 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr5_-_27038683 | 0.32 |
ENST00000511822.1 ENST00000231021.4 |
CDH9 |
cadherin 9, type 2 (T1-cadherin) |
| chr6_+_3118926 | 0.32 |
ENST00000380379.5 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
| chr12_-_57030115 | 0.31 |
ENST00000379441.3 ENST00000179765.5 ENST00000551812.1 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
| chr17_+_4613918 | 0.31 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chr11_-_31832581 | 0.30 |
ENST00000379111.2 |
PAX6 |
paired box 6 |
| chr6_+_143929307 | 0.30 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr19_-_57988871 | 0.30 |
ENST00000596831.1 ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9 ZNF772 |
Uncharacterized protein zinc finger protein 772 |
| chr1_+_215747118 | 0.30 |
ENST00000448333.1 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
| chr1_+_57320437 | 0.29 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr20_-_23066953 | 0.29 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr2_-_136743169 | 0.29 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr18_+_32621324 | 0.29 |
ENST00000300249.5 ENST00000538170.2 ENST00000588910.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr6_+_33168597 | 0.29 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr19_-_48894104 | 0.28 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr6_+_33168637 | 0.28 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr1_+_150898812 | 0.28 |
ENST00000271640.5 ENST00000448029.1 ENST00000368962.2 ENST00000534805.1 ENST00000368969.4 ENST00000368963.1 ENST00000498193.1 |
SETDB1 |
SET domain, bifurcated 1 |
| chr3_-_48471454 | 0.27 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr8_-_6420777 | 0.27 |
ENST00000415216.1 |
ANGPT2 |
angiopoietin 2 |
| chr19_+_39881951 | 0.27 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr7_+_139529040 | 0.27 |
ENST00000455353.1 ENST00000458722.1 ENST00000411653.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr17_+_43138664 | 0.27 |
ENST00000258960.2 |
NMT1 |
N-myristoyltransferase 1 |
| chr19_+_36208877 | 0.27 |
ENST00000420124.1 ENST00000222270.7 ENST00000341701.1 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
| chr20_+_36405665 | 0.27 |
ENST00000373469.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr7_+_139528952 | 0.26 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr20_+_60718785 | 0.26 |
ENST00000421564.1 ENST00000450482.1 ENST00000331758.3 |
SS18L1 |
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr10_+_103892787 | 0.25 |
ENST00000278070.2 ENST00000413464.2 |
PPRC1 |
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr8_-_6420930 | 0.24 |
ENST00000325203.5 |
ANGPT2 |
angiopoietin 2 |
| chr6_-_33168391 | 0.24 |
ENST00000374685.4 ENST00000413614.2 ENST00000374680.3 |
RXRB |
retinoid X receptor, beta |
| chr12_-_91451758 | 0.23 |
ENST00000266719.3 |
KERA |
keratocan |
| chr14_-_69261310 | 0.22 |
ENST00000336440.3 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr2_+_210517895 | 0.22 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr7_+_145813453 | 0.21 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr11_-_62414070 | 0.21 |
ENST00000540933.1 ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB |
glucosidase, alpha; neutral AB |
| chr10_-_65028817 | 0.20 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr8_+_31497271 | 0.20 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr1_+_154540246 | 0.20 |
ENST00000368476.3 |
CHRNB2 |
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr17_-_39646116 | 0.20 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr4_-_87281224 | 0.20 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr17_-_46667628 | 0.19 |
ENST00000498678.1 |
HOXB3 |
homeobox B3 |
| chr8_+_85618155 | 0.18 |
ENST00000523850.1 ENST00000521376.1 |
RALYL |
RALY RNA binding protein-like |
| chrX_+_49644470 | 0.18 |
ENST00000508866.2 |
USP27X |
ubiquitin specific peptidase 27, X-linked |
| chr2_-_183731882 | 0.17 |
ENST00000295113.4 |
FRZB |
frizzled-related protein |
| chr17_-_46667594 | 0.17 |
ENST00000476342.1 ENST00000460160.1 ENST00000472863.1 |
HOXB3 |
homeobox B3 |
| chr8_-_6420759 | 0.17 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr5_+_66300446 | 0.17 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_122085469 | 0.16 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr15_+_78632666 | 0.16 |
ENST00000299529.6 |
CRABP1 |
cellular retinoic acid binding protein 1 |
| chr2_-_136743039 | 0.16 |
ENST00000537273.1 |
DARS |
aspartyl-tRNA synthetase |
| chr11_+_92085707 | 0.16 |
ENST00000525166.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr1_-_40367530 | 0.16 |
ENST00000372816.2 ENST00000372815.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_+_206557366 | 0.16 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_-_114343039 | 0.16 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_-_6420565 | 0.15 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr22_+_31742875 | 0.15 |
ENST00000504184.2 |
AC005003.1 |
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr11_-_30038490 | 0.15 |
ENST00000328224.6 |
KCNA4 |
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr2_-_211036051 | 0.14 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr1_+_87797351 | 0.14 |
ENST00000370542.1 |
LMO4 |
LIM domain only 4 |
| chr2_-_111291587 | 0.14 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr2_+_234601512 | 0.14 |
ENST00000305139.6 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_+_161494770 | 0.14 |
ENST00000414552.2 ENST00000361925.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr11_+_394196 | 0.14 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr22_+_41487711 | 0.13 |
ENST00000263253.7 |
EP300 |
E1A binding protein p300 |
| chr14_+_21785693 | 0.13 |
ENST00000382933.4 ENST00000557351.1 |
RPGRIP1 |
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr3_-_47950745 | 0.12 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr9_-_139371533 | 0.12 |
ENST00000290037.6 ENST00000431893.2 ENST00000371706.3 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chrY_+_4868267 | 0.12 |
ENST00000333703.4 |
PCDH11Y |
protocadherin 11 Y-linked |
| chr9_+_116263778 | 0.12 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr9_-_139372141 | 0.12 |
ENST00000313050.7 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chrX_-_11308598 | 0.12 |
ENST00000380717.3 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr11_+_94706804 | 0.11 |
ENST00000335080.5 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr5_+_161495038 | 0.11 |
ENST00000393933.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_-_99548524 | 0.11 |
ENST00000549558.2 ENST00000550693.2 ENST00000549493.2 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_+_24553933 | 0.11 |
ENST00000424834.2 ENST00000439928.2 |
SPATA13 RP11-309I15.1 |
spermatogenesis associated 13 RP11-309I15.1 |
| chr4_-_74088800 | 0.11 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr4_+_75230853 | 0.10 |
ENST00000244869.2 |
EREG |
epiregulin |
| chr4_+_88754113 | 0.10 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr8_+_85095769 | 0.09 |
ENST00000518566.1 |
RALYL |
RALY RNA binding protein-like |
| chr18_+_18943554 | 0.09 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chrX_+_16141667 | 0.09 |
ENST00000380289.2 |
GRPR |
gastrin-releasing peptide receptor |
| chr8_+_107738240 | 0.09 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr7_+_148892557 | 0.09 |
ENST00000262085.3 |
ZNF282 |
zinc finger protein 282 |
| chr4_+_160188889 | 0.08 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr4_-_87281196 | 0.08 |
ENST00000359221.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr8_+_134125727 | 0.08 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr9_-_13165457 | 0.08 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chrY_+_23698778 | 0.08 |
ENST00000303902.5 |
RBMY1A1 |
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr4_-_76555657 | 0.08 |
ENST00000307465.4 |
CDKL2 |
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr3_+_155838337 | 0.07 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chrX_+_91034260 | 0.07 |
ENST00000395337.2 |
PCDH11X |
protocadherin 11 X-linked |
| chr2_+_210444748 | 0.07 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr1_-_227505289 | 0.07 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr16_+_29789561 | 0.07 |
ENST00000400752.4 |
ZG16 |
zymogen granule protein 16 |
| chr10_+_102222798 | 0.07 |
ENST00000343737.5 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
| chr19_+_19496624 | 0.07 |
ENST00000494516.2 ENST00000360315.3 ENST00000252577.5 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr10_+_21823243 | 0.07 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr6_-_56819385 | 0.07 |
ENST00000370754.5 ENST00000449297.2 |
DST |
dystonin |
| chrX_-_151619746 | 0.06 |
ENST00000370314.4 |
GABRA3 |
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr6_+_31514622 | 0.06 |
ENST00000376146.4 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr4_-_116034979 | 0.06 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr9_-_95186739 | 0.05 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr6_+_45296391 | 0.05 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr5_+_161494521 | 0.05 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_-_173176452 | 0.05 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr12_-_6483969 | 0.05 |
ENST00000396966.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr19_-_39881669 | 0.05 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr16_+_30675654 | 0.05 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr4_+_158142750 | 0.04 |
ENST00000505888.1 ENST00000449365.1 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
| chr7_+_111846741 | 0.04 |
ENST00000421043.1 ENST00000425229.1 ENST00000450657.1 |
ZNF277 |
zinc finger protein 277 |
| chr17_+_2240916 | 0.04 |
ENST00000574563.1 |
SGSM2 |
small G protein signaling modulator 2 |
| chr1_+_93297622 | 0.04 |
ENST00000315741.5 |
RPL5 |
ribosomal protein L5 |
| chr8_+_26247878 | 0.04 |
ENST00000518611.1 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr10_-_65028938 | 0.04 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr2_+_210444142 | 0.04 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr11_-_84634217 | 0.03 |
ENST00000524982.1 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr2_-_60780536 | 0.03 |
ENST00000538214.1 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr3_-_114343768 | 0.03 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_+_85095553 | 0.03 |
ENST00000521268.1 |
RALYL |
RALY RNA binding protein-like |
| chr20_-_18447667 | 0.02 |
ENST00000262547.5 ENST00000329494.5 ENST00000357236.4 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
| chr13_-_114843416 | 0.01 |
ENST00000389544.4 |
RASA3 |
RAS p21 protein activator 3 |
| chr7_-_135194822 | 0.01 |
ENST00000428680.2 ENST00000315544.5 ENST00000423368.2 ENST00000451834.1 ENST00000361528.4 ENST00000356162.4 ENST00000541284.1 |
CNOT4 |
CCR4-NOT transcription complex, subunit 4 |
| chr3_-_113897899 | 0.01 |
ENST00000383673.2 ENST00000295881.7 |
DRD3 |
dopamine receptor D3 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 4.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 2.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 2.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 4.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.9 | 2.7 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.8 | 2.3 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.7 | 2.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.7 | 2.0 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.5 | 2.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.5 | 2.8 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 3.0 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.3 | 1.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 1.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 1.6 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.3 | 2.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.6 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.8 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.2 | 0.6 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 1.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.2 | 0.5 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.6 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.6 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.4 | GO:0052314 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 1.8 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.4 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.3 | GO:0018201 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.4 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 1.8 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.8 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 1.0 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.5 | GO:1905247 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.1 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 2.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 1.0 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 2.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.4 | 2.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 0.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.2 | 2.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 2.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.5 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.2 | 0.7 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.2 | 4.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 4.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.3 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 1.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 2.1 | GO:0070405 | ammonium ion binding(GO:0070405) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |