Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
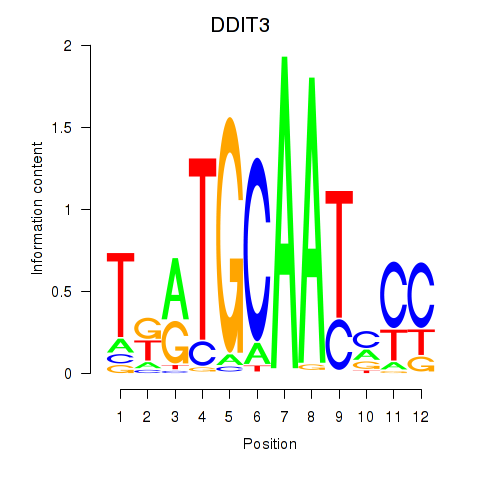
Results for DDIT3
Z-value: 0.63
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DDIT3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DDIT3 | hg19_v2_chr12_-_57914275_57914304 | -0.24 | 3.8e-01 | Click! |
Activity profile of DDIT3 motif
Sorted Z-values of DDIT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DDIT3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_216300784 | 3.22 |
ENST00000421182.1 ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1 |
fibronectin 1 |
| chr3_+_157154578 | 2.91 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr10_-_17659357 | 1.88 |
ENST00000326961.6 ENST00000361271.3 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr2_-_161350305 | 1.87 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr10_-_92681033 | 1.75 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr4_-_99578789 | 1.53 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr2_+_192141611 | 1.44 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr10_-_17659234 | 1.36 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_-_18744528 | 1.26 |
ENST00000523619.1 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr9_+_112542591 | 0.96 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr2_-_179672142 | 0.94 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr9_+_112542572 | 0.93 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr3_+_38017264 | 0.81 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr12_-_52867569 | 0.79 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr5_-_54281407 | 0.75 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr16_+_56965960 | 0.70 |
ENST00000439977.2 ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr15_+_85923797 | 0.69 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr1_+_169079823 | 0.61 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr2_-_208030647 | 0.57 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr4_+_157997273 | 0.54 |
ENST00000541722.1 ENST00000512619.1 |
GLRB |
glycine receptor, beta |
| chr16_+_56970567 | 0.53 |
ENST00000563911.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr10_-_116444371 | 0.53 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr4_+_157997209 | 0.48 |
ENST00000264428.4 |
GLRB |
glycine receptor, beta |
| chr17_+_4855053 | 0.43 |
ENST00000518175.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr7_-_56101826 | 0.41 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr3_-_45883558 | 0.38 |
ENST00000445698.1 ENST00000296135.6 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr15_+_41245160 | 0.37 |
ENST00000444189.2 ENST00000446533.3 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr8_-_93029865 | 0.36 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_+_56315936 | 0.35 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr17_+_56315787 | 0.27 |
ENST00000262290.4 ENST00000421678.2 |
LPO |
lactoperoxidase |
| chr7_-_44530479 | 0.25 |
ENST00000355451.7 |
NUDCD3 |
NudC domain containing 3 |
| chr20_+_24929866 | 0.23 |
ENST00000480798.1 ENST00000376835.2 |
CST7 |
cystatin F (leukocystatin) |
| chr5_-_137878887 | 0.22 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr12_-_53074182 | 0.22 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr11_+_62495541 | 0.20 |
ENST00000530625.1 ENST00000513247.2 |
TTC9C |
tetratricopeptide repeat domain 9C |
| chr2_-_32390801 | 0.19 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr20_+_48807351 | 0.19 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr13_-_24007815 | 0.17 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr16_-_68269971 | 0.17 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr14_-_92413727 | 0.16 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr14_-_92413353 | 0.16 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr3_+_15643476 | 0.16 |
ENST00000436193.1 ENST00000383778.4 |
BTD |
biotinidase |
| chr6_+_31895254 | 0.15 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr15_+_85923856 | 0.12 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr3_+_186330712 | 0.12 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr10_+_86004802 | 0.09 |
ENST00000359452.4 ENST00000358110.5 ENST00000372092.3 |
RGR |
retinal G protein coupled receptor |
| chr1_+_150954493 | 0.08 |
ENST00000368947.4 |
ANXA9 |
annexin A9 |
| chr14_+_21387508 | 0.08 |
ENST00000555624.1 |
RP11-84C10.2 |
RP11-84C10.2 |
| chr5_+_72143988 | 0.07 |
ENST00000506351.2 |
TNPO1 |
transportin 1 |
| chrX_-_40506766 | 0.07 |
ENST00000378421.1 ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38 |
chromosome X open reading frame 38 |
| chr3_+_130569429 | 0.06 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr17_-_34257731 | 0.06 |
ENST00000431884.2 ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1 |
RAD52 motif 1 |
| chr6_-_157744531 | 0.05 |
ENST00000400788.4 ENST00000367144.4 |
TMEM242 |
transmembrane protein 242 |
| chr1_-_161277210 | 0.05 |
ENST00000491222.2 |
MPZ |
myelin protein zero |
| chr9_-_95056010 | 0.05 |
ENST00000443024.2 |
IARS |
isoleucyl-tRNA synthetase |
| chr6_+_168399772 | 0.04 |
ENST00000443060.2 |
KIF25 |
kinesin family member 25 |
| chr22_-_30901637 | 0.04 |
ENST00000381982.3 ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4 |
SEC14-like 4 (S. cerevisiae) |
| chr17_-_46691990 | 0.04 |
ENST00000576562.1 |
HOXB8 |
homeobox B8 |
| chr4_-_2935674 | 0.04 |
ENST00000514800.1 |
MFSD10 |
major facilitator superfamily domain containing 10 |
| chr11_+_61976137 | 0.03 |
ENST00000244930.4 |
SCGB2A1 |
secretoglobin, family 2A, member 1 |
| chr12_-_25102252 | 0.03 |
ENST00000261192.7 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr4_-_119274121 | 0.02 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr2_-_63815628 | 0.01 |
ENST00000409562.3 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chrX_-_100129128 | 0.01 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
| chr8_-_21669826 | 0.01 |
ENST00000517328.1 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr7_-_99006443 | 0.00 |
ENST00000350498.3 |
PDAP1 |
PDGFA associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.2 | 3.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.6 | 2.9 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.3 | 1.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.3 | 0.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 0.8 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 0.6 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.8 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 1.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.4 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 1.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.8 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 1.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 2.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 0.8 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 1.0 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |