Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
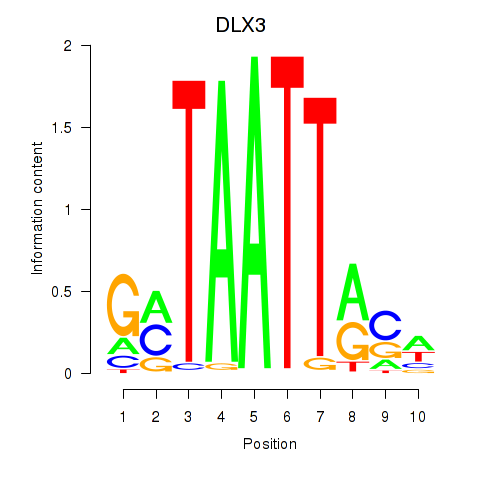
Results for DLX3_EVX1_MEOX1
Z-value: 1.00
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | DLX3 |
|
EVX1
|
ENSG00000106038.8 | EVX1 |
|
MEOX1
|
ENSG00000005102.8 | MEOX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EVX1 | hg19_v2_chr7_+_27282319_27282394 | -0.23 | 3.9e-01 | Click! |
| MEOX1 | hg19_v2_chr17_-_41738931_41739040, hg19_v2_chr17_-_41739283_41739322 | -0.05 | 8.5e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_55562479 | 4.41 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55563072 | 4.18 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_55562582 | 3.62 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr15_-_37393406 | 2.54 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr12_-_6233828 | 2.34 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr11_-_33913708 | 2.29 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_32157947 | 2.25 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr16_-_29910853 | 1.61 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr13_-_46716969 | 1.30 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_+_75080883 | 1.27 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr9_+_116225999 | 1.17 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr7_+_116660246 | 1.15 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr13_-_36788718 | 1.09 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr18_+_32556892 | 0.87 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_99279928 | 0.85 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr3_+_44840679 | 0.79 |
ENST00000425755.1 |
KIF15 |
kinesin family member 15 |
| chr7_-_99717463 | 0.79 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_14029283 | 0.71 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr13_+_58206655 | 0.68 |
ENST00000377918.3 |
PCDH17 |
protocadherin 17 |
| chr5_-_141249154 | 0.67 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr8_-_17533838 | 0.59 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr2_-_61697862 | 0.58 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr8_-_82395461 | 0.58 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr4_+_74275057 | 0.57 |
ENST00000511370.1 |
ALB |
albumin |
| chr6_-_76203345 | 0.56 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr8_+_40010989 | 0.56 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chr6_+_151646800 | 0.54 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr17_-_64225508 | 0.54 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_56694142 | 0.52 |
ENST00000550655.1 ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS |
citrate synthase |
| chr1_-_190446759 | 0.50 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr7_-_87342564 | 0.50 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr3_-_141747950 | 0.50 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_196313239 | 0.49 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr21_-_35899113 | 0.49 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr4_-_174451370 | 0.47 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr6_-_31107127 | 0.46 |
ENST00000259845.4 |
PSORS1C2 |
psoriasis susceptibility 1 candidate 2 |
| chr1_-_68698197 | 0.46 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr8_+_50824233 | 0.45 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr1_+_214161272 | 0.44 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr1_+_198608146 | 0.42 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr1_+_111415757 | 0.42 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr7_-_100860851 | 0.42 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr4_-_116034979 | 0.42 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr4_-_66536057 | 0.41 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr4_-_155533787 | 0.41 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr15_-_64673630 | 0.41 |
ENST00000558008.1 ENST00000559519.1 ENST00000380258.2 |
KIAA0101 |
KIAA0101 |
| chr11_-_107729887 | 0.40 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr2_-_17981462 | 0.39 |
ENST00000402989.1 ENST00000428868.1 |
SMC6 |
structural maintenance of chromosomes 6 |
| chr15_-_65903407 | 0.39 |
ENST00000395644.4 ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9 |
von Willebrand factor A domain containing 9 |
| chr4_+_40198527 | 0.38 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr3_-_33686743 | 0.38 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr4_-_66536196 | 0.38 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr6_+_135502501 | 0.37 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_-_27169801 | 0.36 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chrX_-_118986911 | 0.35 |
ENST00000276201.2 ENST00000345865.2 |
UPF3B |
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr17_-_46690839 | 0.35 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr15_+_96869165 | 0.35 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr5_-_159827033 | 0.34 |
ENST00000523213.1 |
C5orf54 |
chromosome 5 open reading frame 54 |
| chr15_-_64673665 | 0.34 |
ENST00000300035.4 |
KIAA0101 |
KIAA0101 |
| chr12_+_28410128 | 0.34 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr11_+_128563652 | 0.33 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_214161854 | 0.33 |
ENST00000435016.1 |
PROX1 |
prospero homeobox 1 |
| chr17_+_42264556 | 0.33 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr17_-_48785216 | 0.32 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr4_-_105416039 | 0.31 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr18_+_55888767 | 0.31 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_-_111804905 | 0.30 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr13_-_31191642 | 0.30 |
ENST00000405805.1 |
HMGB1 |
high mobility group box 1 |
| chr3_-_178984759 | 0.29 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr6_+_34204642 | 0.28 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr15_+_89631647 | 0.27 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr7_-_14029515 | 0.26 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr10_+_35484793 | 0.26 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr6_+_31895254 | 0.26 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr15_+_58702742 | 0.26 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr1_-_197115818 | 0.26 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_+_169757750 | 0.26 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr12_-_719573 | 0.26 |
ENST00000397265.3 |
NINJ2 |
ninjurin 2 |
| chr7_-_73038867 | 0.25 |
ENST00000313375.3 ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL |
MLX interacting protein-like |
| chr13_-_41593425 | 0.25 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr16_+_69345243 | 0.25 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr17_-_33446820 | 0.25 |
ENST00000592577.1 ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D |
RAD51 paralog D |
| chr4_+_86396265 | 0.25 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_-_55647057 | 0.25 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr9_+_125133315 | 0.24 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_71182695 | 0.24 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr8_-_42234745 | 0.23 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_+_160160346 | 0.23 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_101003687 | 0.23 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr8_+_28748765 | 0.22 |
ENST00000355231.5 |
HMBOX1 |
homeobox containing 1 |
| chr4_-_139163491 | 0.22 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr7_-_81399438 | 0.22 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_73038822 | 0.22 |
ENST00000414749.2 ENST00000429400.2 ENST00000434326.1 |
MLXIPL |
MLX interacting protein-like |
| chr14_+_95027772 | 0.21 |
ENST00000555095.1 ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4 SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr15_-_68497657 | 0.21 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr6_-_33160231 | 0.21 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chr1_-_152386732 | 0.21 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr17_-_33446735 | 0.20 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr14_+_22739823 | 0.20 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr4_-_39979576 | 0.19 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr1_-_151431647 | 0.19 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr5_-_134871639 | 0.19 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr3_+_157828152 | 0.19 |
ENST00000476899.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chr12_-_14133053 | 0.19 |
ENST00000609686.1 |
GRIN2B |
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr6_-_55739542 | 0.19 |
ENST00000446683.2 |
BMP5 |
bone morphogenetic protein 5 |
| chr3_-_108248169 | 0.19 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr2_-_55646957 | 0.19 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr8_-_103425047 | 0.19 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr1_+_236958554 | 0.18 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr5_+_31193847 | 0.18 |
ENST00000514738.1 ENST00000265071.2 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr7_-_25268104 | 0.18 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr5_+_156712372 | 0.18 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_160160283 | 0.18 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr11_-_117747327 | 0.18 |
ENST00000584230.1 ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6 FXYD6-FXYD2 |
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr6_-_87804815 | 0.17 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr2_+_202047596 | 0.17 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr18_+_32173276 | 0.17 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr7_-_99716952 | 0.17 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr5_+_159343688 | 0.17 |
ENST00000306675.3 |
ADRA1B |
adrenoceptor alpha 1B |
| chr20_+_45947246 | 0.16 |
ENST00000599904.1 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr5_-_34043310 | 0.16 |
ENST00000231338.7 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
| chr13_-_95131923 | 0.16 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chr1_-_151431909 | 0.16 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr8_-_103424916 | 0.15 |
ENST00000220959.4 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr14_-_36988882 | 0.15 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr4_+_144354644 | 0.15 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr11_+_20620946 | 0.15 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr14_+_32798547 | 0.14 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr12_+_41831485 | 0.14 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr22_-_30960876 | 0.14 |
ENST00000401975.1 ENST00000428682.1 ENST00000423299.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr10_+_114710211 | 0.14 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr15_+_93443419 | 0.14 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr14_-_51027838 | 0.13 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr8_-_103424986 | 0.13 |
ENST00000521922.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr7_+_73245193 | 0.13 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr2_-_37544209 | 0.13 |
ENST00000234179.2 |
PRKD3 |
protein kinase D3 |
| chr10_+_114710425 | 0.13 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr14_-_21737610 | 0.13 |
ENST00000320084.7 ENST00000449098.1 ENST00000336053.6 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr20_+_30697298 | 0.12 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr14_-_21737551 | 0.12 |
ENST00000554891.1 ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC |
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr3_-_57233966 | 0.12 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr3_-_38071122 | 0.12 |
ENST00000334661.4 |
PLCD1 |
phospholipase C, delta 1 |
| chr6_+_160211481 | 0.12 |
ENST00000367034.4 |
MRPL18 |
mitochondrial ribosomal protein L18 |
| chr7_-_14028488 | 0.12 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr12_-_53171128 | 0.11 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chrX_-_19988382 | 0.11 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr2_+_171785012 | 0.11 |
ENST00000234160.4 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
| chr20_+_42187608 | 0.11 |
ENST00000373100.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr15_+_89164520 | 0.11 |
ENST00000332810.3 |
AEN |
apoptosis enhancing nuclease |
| chr4_-_87028478 | 0.11 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr11_-_70672645 | 0.11 |
ENST00000423696.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr6_-_111136513 | 0.11 |
ENST00000368911.3 |
CDK19 |
cyclin-dependent kinase 19 |
| chr16_-_66864806 | 0.11 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr14_+_22748980 | 0.11 |
ENST00000390465.2 |
TRAV38-2DV8 |
T cell receptor alpha variable 38-2/delta variable 8 |
| chr12_-_118628350 | 0.10 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr10_-_73848086 | 0.10 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr17_-_60142609 | 0.10 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr6_-_109702885 | 0.10 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr18_-_3220106 | 0.10 |
ENST00000356443.4 ENST00000400569.3 |
MYOM1 |
myomesin 1 |
| chr2_-_214016314 | 0.09 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr1_-_101491319 | 0.09 |
ENST00000342173.7 ENST00000488176.1 ENST00000370109.3 |
DPH5 |
diphthamide biosynthesis 5 |
| chr8_+_77593448 | 0.09 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr16_-_67517716 | 0.09 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr13_+_33590553 | 0.09 |
ENST00000380099.3 |
KL |
klotho |
| chr20_+_42187682 | 0.09 |
ENST00000373092.3 ENST00000373077.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr12_-_10978957 | 0.09 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chrX_-_23926004 | 0.09 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr11_-_117748138 | 0.08 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr2_-_86422095 | 0.08 |
ENST00000254636.5 |
IMMT |
inner membrane protein, mitochondrial |
| chr11_-_115630900 | 0.08 |
ENST00000537070.1 ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900 |
long intergenic non-protein coding RNA 900 |
| chr7_+_99425633 | 0.08 |
ENST00000354829.2 ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43 |
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr2_+_202047843 | 0.08 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr7_+_50348268 | 0.08 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr12_-_86650077 | 0.08 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr7_-_81399355 | 0.08 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_171034646 | 0.08 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr7_-_81399329 | 0.08 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr19_-_46088068 | 0.08 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr12_-_121454148 | 0.07 |
ENST00000535367.1 ENST00000538296.1 ENST00000445832.3 ENST00000536407.2 ENST00000366211.2 ENST00000539736.1 ENST00000288757.3 ENST00000537817.1 |
C12orf43 |
chromosome 12 open reading frame 43 |
| chr7_-_81399411 | 0.07 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_+_100485712 | 0.07 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chr9_+_125132803 | 0.07 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_-_36989336 | 0.07 |
ENST00000522719.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr1_-_28384598 | 0.07 |
ENST00000373864.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr6_+_50786414 | 0.07 |
ENST00000344788.3 ENST00000393655.3 ENST00000263046.4 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr2_+_201754050 | 0.07 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr8_+_110346546 | 0.07 |
ENST00000521662.1 ENST00000521688.1 ENST00000520147.1 |
ENY2 |
enhancer of yellow 2 homolog (Drosophila) |
| chrX_+_99839799 | 0.07 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chrX_-_16887963 | 0.07 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr11_+_24518723 | 0.07 |
ENST00000336930.6 ENST00000529015.1 ENST00000533227.1 |
LUZP2 |
leucine zipper protein 2 |
| chr4_-_109684120 | 0.07 |
ENST00000512646.1 ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL |
ethanolamine-phosphate phospho-lyase |
| chr16_+_30675654 | 0.07 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr22_-_41682172 | 0.07 |
ENST00000356244.3 |
RANGAP1 |
Ran GTPase activating protein 1 |
| chr10_-_69455873 | 0.06 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr3_-_113897899 | 0.06 |
ENST00000383673.2 ENST00000295881.7 |
DRD3 |
dopamine receptor D3 |
| chr12_-_86650045 | 0.06 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr4_+_169013666 | 0.06 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr7_-_25019760 | 0.06 |
ENST00000352860.1 ENST00000353930.1 ENST00000431825.2 ENST00000313367.2 |
OSBPL3 |
oxysterol binding protein-like 3 |
| chrX_-_110655306 | 0.06 |
ENST00000371993.2 |
DCX |
doublecortin |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 0.8 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.3 | 1.3 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 0.6 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.4 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 2.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.4 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.7 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.2 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.1 | 0.5 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.1 | 2.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.4 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.3 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.3 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.2 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.1 | 0.5 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.3 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.5 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 2.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.0 | 0.3 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.2 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.6 | GO:0046464 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.0 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 14.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 0.8 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.2 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.5 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.5 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 2.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 1.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.3 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 2.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |