Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
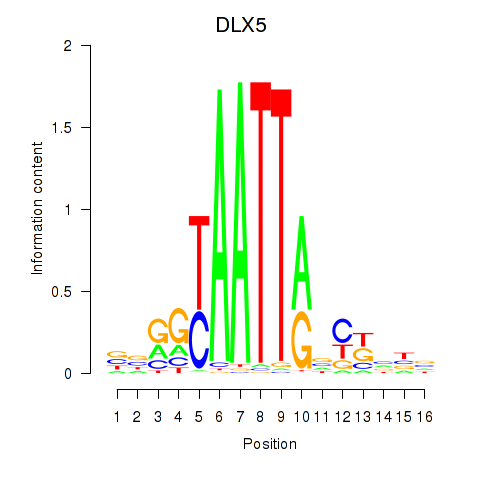
Results for DLX5
Z-value: 0.57
Transcription factors associated with DLX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX5
|
ENSG00000105880.4 | DLX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX5 | hg19_v2_chr7_-_96654133_96654409 | 0.05 | 8.4e-01 | Click! |
Activity profile of DLX5 motif
Sorted Z-values of DLX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_81110684 | 2.00 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr4_-_143767428 | 1.40 |
ENST00000513000.1 ENST00000509777.1 ENST00000503927.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_+_100770328 | 1.12 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr12_+_54378923 | 1.09 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr12_-_50616382 | 0.82 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr8_+_32579341 | 0.72 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr6_-_112194484 | 0.48 |
ENST00000518295.1 ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr2_+_30369859 | 0.48 |
ENST00000402003.3 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr2_+_30369807 | 0.47 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr11_-_102651343 | 0.42 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr1_-_28969517 | 0.39 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr18_-_33709268 | 0.39 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr2_-_220118631 | 0.38 |
ENST00000248437.4 |
TUBA4A |
tubulin, alpha 4a |
| chr19_+_36606354 | 0.32 |
ENST00000589996.1 ENST00000591296.1 |
TBCB |
tubulin folding cofactor B |
| chr19_+_36605850 | 0.31 |
ENST00000221855.3 |
TBCB |
tubulin folding cofactor B |
| chr15_+_89631381 | 0.29 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr11_-_26743546 | 0.28 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr14_-_68283291 | 0.26 |
ENST00000555452.1 ENST00000347230.4 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
| chr14_-_24711470 | 0.23 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr12_+_7014126 | 0.20 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr3_-_105588231 | 0.19 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_7013897 | 0.19 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr12_-_10151773 | 0.19 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr14_-_24711865 | 0.19 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr14_-_24711806 | 0.18 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_202317815 | 0.18 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr11_-_71823715 | 0.17 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr11_-_71823266 | 0.16 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr12_+_7014064 | 0.16 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr1_-_23670817 | 0.13 |
ENST00000478691.1 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr1_-_23670813 | 0.11 |
ENST00000374612.1 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr19_-_14785674 | 0.10 |
ENST00000253673.5 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_+_58570605 | 0.10 |
ENST00000359978.6 ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135 |
zinc finger protein 135 |
| chr3_-_105587879 | 0.10 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr19_-_14785622 | 0.09 |
ENST00000443157.2 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr19_-_14785698 | 0.09 |
ENST00000344373.4 ENST00000595472.1 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr2_-_183387064 | 0.09 |
ENST00000536095.1 ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_8873531 | 0.08 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chr1_-_23670752 | 0.08 |
ENST00000302271.6 ENST00000426846.2 ENST00000427764.2 ENST00000606561.1 ENST00000374616.3 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr20_+_43849941 | 0.08 |
ENST00000372769.3 |
SEMG2 |
semenogelin II |
| chr8_-_22526597 | 0.08 |
ENST00000519513.1 ENST00000276416.6 ENST00000520292.1 ENST00000522268.1 |
BIN3 |
bridging integrator 3 |
| chr6_-_38670897 | 0.08 |
ENST00000373365.4 |
GLO1 |
glyoxalase I |
| chr7_+_97736197 | 0.08 |
ENST00000297293.5 |
LMTK2 |
lemur tyrosine kinase 2 |
| chr12_-_57039739 | 0.07 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr17_-_37009882 | 0.06 |
ENST00000378096.3 ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23 |
ribosomal protein L23 |
| chr9_+_34646651 | 0.05 |
ENST00000378842.3 |
GALT |
galactose-1-phosphate uridylyltransferase |
| chr1_+_101003687 | 0.04 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr12_-_53171128 | 0.04 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr7_-_86849883 | 0.04 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr9_+_34646624 | 0.04 |
ENST00000450095.2 ENST00000556278.1 |
GALT GALT |
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr14_+_24540046 | 0.04 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr2_-_183387283 | 0.03 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_7069760 | 0.03 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr4_+_151503077 | 0.03 |
ENST00000317605.4 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
| chr3_+_11178779 | 0.03 |
ENST00000438284.2 |
HRH1 |
histamine receptor H1 |
| chr1_-_78444738 | 0.02 |
ENST00000436586.2 ENST00000370768.2 |
FUBP1 |
far upstream element (FUSE) binding protein 1 |
| chr17_-_47785504 | 0.02 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr1_+_50569575 | 0.02 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr1_+_160160283 | 0.02 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr7_-_99716952 | 0.02 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_+_158519654 | 0.02 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr14_+_96000930 | 0.01 |
ENST00000331334.4 |
GLRX5 |
glutaredoxin 5 |
| chr13_-_36050819 | 0.01 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr1_+_160160346 | 0.01 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_54569968 | 0.01 |
ENST00000391366.1 |
AL161915.1 |
Uncharacterized protein |
| chr2_+_169757750 | 0.00 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr5_-_160279207 | 0.00 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr19_+_54641444 | 0.00 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 1.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.5 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.8 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.6 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |