Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
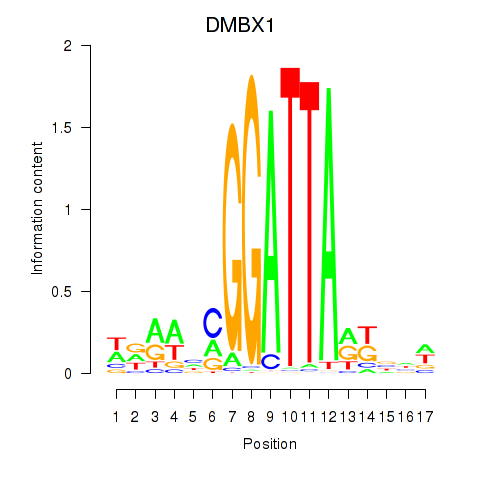
Results for DMBX1
Z-value: 0.50
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | DMBX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg19_v2_chr1_+_46972668_46972669 | -0.12 | 6.6e-01 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_169552748 | 2.06 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_-_16524357 | 1.16 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr9_-_21995300 | 1.15 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr2_-_175711133 | 1.00 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr6_+_89791507 | 0.97 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr9_-_21994344 | 0.94 |
ENST00000530628.2 ENST00000361570.3 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr12_+_53693812 | 0.94 |
ENST00000549488.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr9_-_21995249 | 0.88 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_-_21994597 | 0.78 |
ENST00000579755.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr3_-_121468513 | 0.75 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr3_-_121468602 | 0.68 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr19_+_41284121 | 0.68 |
ENST00000594800.1 ENST00000357052.2 ENST00000602173.1 |
RAB4B |
RAB4B, member RAS oncogene family |
| chrX_-_48931648 | 0.67 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr3_-_11685345 | 0.43 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr15_-_85201779 | 0.34 |
ENST00000360476.3 ENST00000394588.3 |
NMB |
neuromedin B |
| chr2_+_223725652 | 0.34 |
ENST00000357430.3 ENST00000392066.3 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
| chr18_+_29027696 | 0.33 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr2_+_37571845 | 0.31 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr14_+_50234309 | 0.30 |
ENST00000298307.5 |
KLHDC2 |
kelch domain containing 2 |
| chr6_-_111927062 | 0.28 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr3_-_149293990 | 0.23 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr10_-_35104185 | 0.20 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr8_-_110656995 | 0.14 |
ENST00000276646.9 ENST00000533065.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr10_+_74451883 | 0.13 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chr1_-_94586651 | 0.12 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr15_-_61521495 | 0.11 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr3_-_150920979 | 0.11 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr12_+_54718904 | 0.09 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chr1_+_109756523 | 0.09 |
ENST00000234677.2 ENST00000369923.4 |
SARS |
seryl-tRNA synthetase |
| chr5_+_50678921 | 0.08 |
ENST00000230658.7 |
ISL1 |
ISL LIM homeobox 1 |
| chr3_-_149470229 | 0.08 |
ENST00000473414.1 |
COMMD2 |
COMM domain containing 2 |
| chr12_-_10959892 | 0.08 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr2_-_70780572 | 0.07 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr10_+_86004802 | 0.05 |
ENST00000359452.4 ENST00000358110.5 ENST00000372092.3 |
RGR |
retinal G protein coupled receptor |
| chr11_-_36619771 | 0.04 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chr22_-_43411106 | 0.04 |
ENST00000453643.1 ENST00000263246.3 ENST00000337959.4 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_56727487 | 0.03 |
ENST00000548043.1 ENST00000425394.2 |
PAN2 |
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_-_165414414 | 0.03 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr3_+_138340067 | 0.03 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr12_-_56727676 | 0.02 |
ENST00000547572.1 ENST00000257931.5 ENST00000440411.3 |
PAN2 |
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr20_-_43729750 | 0.02 |
ENST00000537075.1 ENST00000306117.1 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr12_-_103310987 | 0.00 |
ENST00000307000.2 |
PAH |
phenylalanine hydroxylase |
| chr17_-_12921270 | 0.00 |
ENST00000578071.1 ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2 |
elaC ribonuclease Z 2 |
| chr1_-_16563641 | 0.00 |
ENST00000375599.3 |
RSG1 |
REM2 and RAB-like small GTPase 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.2 | 1.2 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 2.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 2.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |