Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
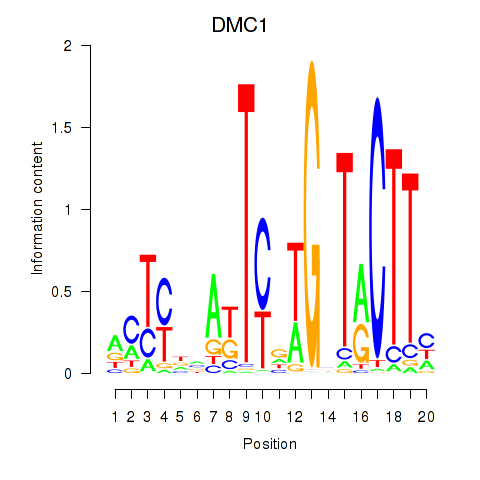
Results for DMC1
Z-value: 0.62
Transcription factors associated with DMC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMC1
|
ENSG00000100206.5 | DMC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMC1 | hg19_v2_chr22_-_38966172_38966291 | 0.63 | 8.9e-03 | Click! |
Activity profile of DMC1 motif
Sorted Z-values of DMC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DMC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_134305322 | 1.54 |
ENST00000276241.6 ENST00000344129.2 |
CXorf48 |
cancer/testis antigen 55 |
| chrX_+_65382433 | 1.44 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr12_-_8803128 | 1.02 |
ENST00000543467.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr3_-_141747950 | 0.97 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_85779971 | 0.85 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_85780086 | 0.73 |
ENST00000532317.1 ENST00000528256.1 ENST00000526033.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr14_-_21491477 | 0.70 |
ENST00000298684.5 ENST00000557169.1 ENST00000553563.1 |
NDRG2 |
NDRG family member 2 |
| chr10_+_11047259 | 0.58 |
ENST00000379261.4 ENST00000416382.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr14_-_21492113 | 0.57 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr5_-_82969405 | 0.53 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr14_-_21492251 | 0.53 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr3_-_141747459 | 0.50 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_+_150758304 | 0.41 |
ENST00000482950.1 ENST00000463414.1 ENST00000310317.5 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr21_-_35281399 | 0.40 |
ENST00000418933.1 |
ATP5O |
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr2_+_233497931 | 0.38 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr7_-_22539771 | 0.37 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr19_-_3600549 | 0.29 |
ENST00000589966.1 |
TBXA2R |
thromboxane A2 receptor |
| chr2_+_219247021 | 0.29 |
ENST00000539932.1 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr2_-_21022818 | 0.27 |
ENST00000440866.2 ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43 |
chromosome 2 open reading frame 43 |
| chr10_-_62332357 | 0.26 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr18_+_56892724 | 0.26 |
ENST00000456142.3 ENST00000530323.1 |
GRP |
gastrin-releasing peptide |
| chrX_+_65382381 | 0.25 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr6_-_150067632 | 0.25 |
ENST00000460354.2 ENST00000367404.4 ENST00000543637.1 |
NUP43 |
nucleoporin 43kDa |
| chr3_+_101546827 | 0.24 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr5_-_135231516 | 0.23 |
ENST00000274520.1 |
IL9 |
interleukin 9 |
| chr1_-_190446759 | 0.23 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr8_+_145438870 | 0.22 |
ENST00000527931.1 |
FAM203B |
family with sequence similarity 203, member B |
| chr1_+_12290121 | 0.21 |
ENST00000358136.3 ENST00000356315.4 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_-_11863571 | 0.20 |
ENST00000376583.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr16_+_2255710 | 0.20 |
ENST00000397124.1 ENST00000565250.1 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr6_-_150067696 | 0.19 |
ENST00000340413.2 ENST00000367403.3 |
NUP43 |
nucleoporin 43kDa |
| chr3_+_148457585 | 0.18 |
ENST00000402260.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr2_+_219246746 | 0.17 |
ENST00000233202.6 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr10_+_17851362 | 0.17 |
ENST00000331429.2 ENST00000457317.1 |
MRC1L1 |
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chrX_-_49965663 | 0.16 |
ENST00000376056.2 ENST00000376058.2 ENST00000358526.2 |
AKAP4 |
A kinase (PRKA) anchor protein 4 |
| chr17_-_60142609 | 0.16 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr19_+_1041212 | 0.14 |
ENST00000433129.1 |
ABCA7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr11_+_71846748 | 0.12 |
ENST00000445078.2 |
FOLR3 |
folate receptor 3 (gamma) |
| chr2_-_48982857 | 0.11 |
ENST00000403273.1 ENST00000401907.1 ENST00000344775.3 |
LHCGR |
luteinizing hormone/choriogonadotropin receptor |
| chr11_+_71846764 | 0.10 |
ENST00000456237.1 ENST00000442948.2 ENST00000546166.1 |
FOLR3 |
folate receptor 3 (gamma) |
| chr10_+_74870206 | 0.09 |
ENST00000357321.4 ENST00000349051.5 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_+_62538248 | 0.08 |
ENST00000448257.2 |
CDK1 |
cyclin-dependent kinase 1 |
| chr2_+_136343820 | 0.08 |
ENST00000410054.1 |
R3HDM1 |
R3H domain containing 1 |
| chr12_-_109025849 | 0.08 |
ENST00000228463.6 |
SELPLG |
selectin P ligand |
| chr16_-_30122717 | 0.08 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr10_+_62538089 | 0.06 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr7_-_15601595 | 0.06 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr10_+_74870253 | 0.06 |
ENST00000544879.1 ENST00000537969.1 ENST00000372997.3 |
NUDT13 |
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr11_+_101785727 | 0.06 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr2_-_163008903 | 0.05 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chrX_-_49965009 | 0.05 |
ENST00000437370.1 ENST00000376064.3 ENST00000448865.1 |
AKAP4 |
A kinase (PRKA) anchor protein 4 |
| chr2_-_48982708 | 0.05 |
ENST00000428232.1 ENST00000405626.1 ENST00000294954.7 |
LHCGR |
luteinizing hormone/choriogonadotropin receptor |
| chr11_+_71238313 | 0.04 |
ENST00000398536.4 |
KRTAP5-7 |
keratin associated protein 5-7 |
| chr18_-_5396271 | 0.04 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr10_-_135187193 | 0.04 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr7_+_47694842 | 0.03 |
ENST00000408988.2 |
C7orf65 |
chromosome 7 open reading frame 65 |
| chr6_-_62996066 | 0.02 |
ENST00000281156.4 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
| chr11_-_73720122 | 0.01 |
ENST00000426995.2 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.5 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 1.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.3 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 1.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 0.5 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.1 | 1.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.3 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 2.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.2 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.0 | 0.1 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 1.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |