Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for DPRX
Z-value: 0.99
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | DPRX |
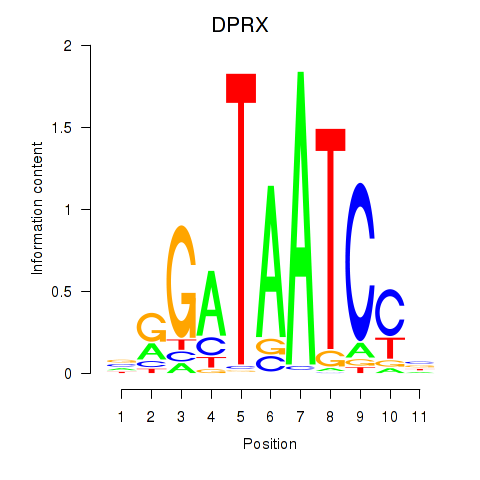
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_321340 | 2.80 |
ENST00000526811.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr3_-_158390282 | 1.79 |
ENST00000264265.3 |
LXN |
latexin |
| chr7_+_150264365 | 1.67 |
ENST00000255945.2 ENST00000461940.1 |
GIMAP4 |
GTPase, IMAP family member 4 |
| chr7_-_15726296 | 1.38 |
ENST00000262041.5 |
MEOX2 |
mesenchyme homeobox 2 |
| chr11_-_104817919 | 1.32 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_33701286 | 1.24 |
ENST00000403687.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_+_54764346 | 1.14 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr20_+_33759854 | 1.03 |
ENST00000216968.4 |
PROCR |
protein C receptor, endothelial |
| chr1_+_169079823 | 1.03 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_-_150780757 | 0.90 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr12_+_113344582 | 0.86 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_321050 | 0.77 |
ENST00000399808.4 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr1_+_81771806 | 0.76 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr1_-_151965048 | 0.69 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr2_+_33661382 | 0.67 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr9_+_116207007 | 0.65 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr12_+_113344755 | 0.60 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_235405168 | 0.59 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chrX_+_99899180 | 0.59 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chrX_+_15525426 | 0.56 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chrX_+_100333709 | 0.56 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr9_+_116225999 | 0.55 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr18_-_52989525 | 0.54 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr12_+_58138664 | 0.54 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr19_+_41284121 | 0.51 |
ENST00000594800.1 ENST00000357052.2 ENST00000602173.1 |
RAB4B |
RAB4B, member RAS oncogene family |
| chr6_-_13487784 | 0.51 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr18_-_52989217 | 0.51 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr14_+_61995722 | 0.50 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr9_-_13279563 | 0.49 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr14_+_91581011 | 0.49 |
ENST00000523894.1 ENST00000522322.1 ENST00000523771.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr10_+_97515409 | 0.48 |
ENST00000371207.3 ENST00000543964.1 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr1_-_173174681 | 0.47 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr12_-_9913489 | 0.46 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr11_+_12399071 | 0.45 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr7_+_134464414 | 0.45 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr8_-_80993010 | 0.44 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr6_+_26383404 | 0.44 |
ENST00000416795.2 ENST00000494184.1 |
BTN2A2 |
butyrophilin, subfamily 2, member A2 |
| chr1_+_16767167 | 0.41 |
ENST00000337132.5 |
NECAP2 |
NECAP endocytosis associated 2 |
| chr3_-_18466787 | 0.41 |
ENST00000338745.6 ENST00000450898.1 |
SATB1 |
SATB homeobox 1 |
| chr11_-_104972158 | 0.40 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr3_-_18466026 | 0.40 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr3_+_158991025 | 0.39 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr1_-_22263790 | 0.39 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr10_+_104178946 | 0.39 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr12_+_58138800 | 0.39 |
ENST00000547992.1 ENST00000552816.1 ENST00000547472.1 |
TSPAN31 |
tetraspanin 31 |
| chr3_+_10068095 | 0.38 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr14_+_85996471 | 0.37 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_16767195 | 0.37 |
ENST00000504551.2 ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2 |
NECAP endocytosis associated 2 |
| chr19_+_2476116 | 0.36 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr4_+_89299885 | 0.36 |
ENST00000380265.5 ENST00000273960.3 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr12_+_113344811 | 0.35 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_104905840 | 0.34 |
ENST00000526568.1 ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
| chr20_+_8112824 | 0.33 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr8_+_1772132 | 0.33 |
ENST00000349830.3 ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr6_-_32908792 | 0.32 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr18_+_56530136 | 0.32 |
ENST00000591083.1 |
ZNF532 |
zinc finger protein 532 |
| chr17_-_15244894 | 0.29 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr7_+_134464376 | 0.29 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr16_+_67562702 | 0.28 |
ENST00000379312.3 ENST00000042381.4 ENST00000540839.3 |
FAM65A |
family with sequence similarity 65, member A |
| chr10_+_74451883 | 0.28 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chrX_-_77150985 | 0.28 |
ENST00000358075.6 |
MAGT1 |
magnesium transporter 1 |
| chr3_+_130650738 | 0.28 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr7_-_94285511 | 0.26 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr3_+_93781728 | 0.25 |
ENST00000314622.4 |
NSUN3 |
NOP2/Sun domain family, member 3 |
| chr1_-_27481401 | 0.25 |
ENST00000263980.3 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
| chr14_+_85996507 | 0.25 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr9_+_17135016 | 0.25 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr8_-_120605194 | 0.23 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr19_-_50370799 | 0.23 |
ENST00000600910.1 ENST00000322344.3 ENST00000600573.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chrX_+_100878079 | 0.21 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr21_-_43346790 | 0.21 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr6_+_26383318 | 0.21 |
ENST00000469230.1 ENST00000490025.1 ENST00000356709.4 ENST00000352867.2 ENST00000493275.1 ENST00000472507.1 ENST00000482536.1 ENST00000432533.2 ENST00000482842.1 |
BTN2A2 |
butyrophilin, subfamily 2, member A2 |
| chr9_+_17134980 | 0.20 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr9_+_2015335 | 0.20 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_93772326 | 0.20 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr14_-_106518922 | 0.19 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr2_-_98280383 | 0.19 |
ENST00000289228.5 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr17_+_8191815 | 0.18 |
ENST00000226105.6 ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF |
RAN guanine nucleotide release factor |
| chrX_+_46771711 | 0.18 |
ENST00000424392.1 ENST00000397189.1 |
PHF16 |
jade family PHD finger 3 |
| chr17_-_19771216 | 0.17 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr7_+_134576151 | 0.17 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr10_-_103578162 | 0.17 |
ENST00000361464.3 ENST00000357797.5 ENST00000370094.3 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chr15_+_63481668 | 0.17 |
ENST00000321437.4 ENST00000559006.1 ENST00000448330.2 |
RAB8B |
RAB8B, member RAS oncogene family |
| chr16_-_21416640 | 0.17 |
ENST00000542817.1 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr10_-_103578182 | 0.17 |
ENST00000439817.1 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chr17_+_73663402 | 0.16 |
ENST00000355423.3 |
SAP30BP |
SAP30 binding protein |
| chr22_+_23161491 | 0.15 |
ENST00000390316.2 |
IGLV3-9 |
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr12_+_56552128 | 0.15 |
ENST00000548580.1 ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr12_+_93772402 | 0.15 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr16_-_21849091 | 0.14 |
ENST00000537951.1 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr10_-_105845674 | 0.14 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr11_-_77123065 | 0.14 |
ENST00000530617.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr17_-_19771242 | 0.14 |
ENST00000361658.2 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr4_-_73935409 | 0.13 |
ENST00000507544.2 ENST00000295890.4 |
COX18 |
COX18 cytochrome C oxidase assembly factor |
| chr18_-_64271363 | 0.13 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr2_-_85555355 | 0.13 |
ENST00000282120.2 ENST00000398263.2 |
TGOLN2 |
trans-golgi network protein 2 |
| chr17_+_77021702 | 0.12 |
ENST00000392445.2 ENST00000354124.3 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr6_-_32784687 | 0.12 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr7_-_97881429 | 0.12 |
ENST00000420697.1 ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
| chr11_-_61560053 | 0.12 |
ENST00000537328.1 |
TMEM258 |
transmembrane protein 258 |
| chr8_+_94752349 | 0.12 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr9_+_35792151 | 0.11 |
ENST00000342694.2 |
NPR2 |
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr14_+_75469606 | 0.11 |
ENST00000266126.5 |
EIF2B2 |
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr13_+_28712614 | 0.11 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr9_-_95244781 | 0.11 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr11_-_77122928 | 0.11 |
ENST00000528203.1 ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_+_64949158 | 0.10 |
ENST00000527739.1 ENST00000526966.1 ENST00000533129.1 ENST00000524773.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr6_-_35480640 | 0.10 |
ENST00000428978.1 ENST00000322263.4 |
TULP1 |
tubby like protein 1 |
| chr8_-_79717750 | 0.10 |
ENST00000263851.4 ENST00000379113.2 |
IL7 |
interleukin 7 |
| chr1_-_9131776 | 0.10 |
ENST00000484798.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr7_+_89975979 | 0.10 |
ENST00000257659.8 ENST00000222511.6 ENST00000417207.1 |
GTPBP10 |
GTP-binding protein 10 (putative) |
| chr19_+_41222998 | 0.10 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr7_+_134576317 | 0.10 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr4_-_87374283 | 0.10 |
ENST00000361569.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr21_+_19617140 | 0.10 |
ENST00000299295.2 ENST00000338326.3 |
CHODL |
chondrolectin |
| chr1_-_94586651 | 0.09 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr4_-_122791583 | 0.09 |
ENST00000506636.1 ENST00000264499.4 |
BBS7 |
Bardet-Biedl syndrome 7 |
| chr9_-_112083229 | 0.09 |
ENST00000374566.3 ENST00000374557.4 |
EPB41L4B |
erythrocyte membrane protein band 4.1 like 4B |
| chr15_+_81475047 | 0.09 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr19_+_36132631 | 0.09 |
ENST00000379026.2 ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2 |
ets variant 2 |
| chr2_-_30144432 | 0.09 |
ENST00000389048.3 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
| chr11_+_64948665 | 0.09 |
ENST00000533820.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr9_-_35096545 | 0.09 |
ENST00000378617.3 ENST00000341666.3 ENST00000361778.2 |
PIGO |
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr1_+_74701062 | 0.09 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr11_-_61560254 | 0.08 |
ENST00000543510.1 |
TMEM258 |
transmembrane protein 258 |
| chr2_+_113342163 | 0.08 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr6_-_32908765 | 0.08 |
ENST00000416244.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr11_+_2323236 | 0.07 |
ENST00000182290.4 |
TSPAN32 |
tetraspanin 32 |
| chr5_-_37835929 | 0.07 |
ENST00000381826.4 ENST00000427982.1 |
GDNF |
glial cell derived neurotrophic factor |
| chr5_-_66492562 | 0.07 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr4_-_47465666 | 0.07 |
ENST00000381571.4 |
COMMD8 |
COMM domain containing 8 |
| chr17_-_42019836 | 0.07 |
ENST00000225992.3 |
PPY |
pancreatic polypeptide |
| chr9_-_123812542 | 0.07 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr16_-_46723066 | 0.07 |
ENST00000299138.7 |
VPS35 |
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr1_-_12891264 | 0.07 |
ENST00000535591.1 ENST00000437584.1 |
PRAMEF11 |
PRAME family member 11 |
| chr6_+_46761118 | 0.07 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr19_+_8455077 | 0.07 |
ENST00000328024.6 |
RAB11B |
RAB11B, member RAS oncogene family |
| chr11_+_94706973 | 0.07 |
ENST00000536741.1 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr21_-_28217721 | 0.06 |
ENST00000284984.3 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr1_+_12916941 | 0.06 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr9_-_13279406 | 0.06 |
ENST00000546205.1 |
MPDZ |
multiple PDZ domain protein |
| chr1_-_45805607 | 0.06 |
ENST00000372104.1 ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH |
mutY homolog |
| chr11_+_60681346 | 0.06 |
ENST00000227525.3 |
TMEM109 |
transmembrane protein 109 |
| chr12_-_7848364 | 0.06 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr7_+_94536898 | 0.06 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr6_+_37225540 | 0.06 |
ENST00000373491.3 |
TBC1D22B |
TBC1 domain family, member 22B |
| chr13_+_52436111 | 0.06 |
ENST00000242819.4 |
CCDC70 |
coiled-coil domain containing 70 |
| chr11_+_64949343 | 0.06 |
ENST00000279247.6 ENST00000532285.1 ENST00000534373.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr19_-_50529193 | 0.05 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr3_+_49058444 | 0.05 |
ENST00000326925.6 ENST00000395458.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr5_-_9712312 | 0.05 |
ENST00000506620.1 ENST00000514078.1 ENST00000606744.1 |
TAS2R1 CTD-2143L24.1 |
taste receptor, type 2, member 1 CTD-2143L24.1 |
| chr12_+_122516626 | 0.05 |
ENST00000319080.7 |
MLXIP |
MLX interacting protein |
| chrX_+_115567767 | 0.05 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr12_+_130554803 | 0.05 |
ENST00000535487.1 |
RP11-474D1.2 |
RP11-474D1.2 |
| chr17_-_3182268 | 0.05 |
ENST00000408891.2 |
OR3A2 |
olfactory receptor, family 3, subfamily A, member 2 |
| chr1_+_36772691 | 0.05 |
ENST00000312808.4 ENST00000505871.1 |
SH3D21 |
SH3 domain containing 21 |
| chr5_-_150948414 | 0.05 |
ENST00000261800.5 |
FAT2 |
FAT atypical cadherin 2 |
| chr7_+_28452130 | 0.05 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr18_-_32957260 | 0.05 |
ENST00000587422.1 ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396 |
zinc finger protein 396 |
| chr19_+_49999631 | 0.05 |
ENST00000270625.2 ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11 |
ribosomal protein S11 |
| chr3_-_124653579 | 0.05 |
ENST00000478191.1 ENST00000311075.3 |
MUC13 |
mucin 13, cell surface associated |
| chr5_+_139781445 | 0.04 |
ENST00000532219.1 ENST00000394722.3 |
ANKHD1-EIF4EBP3 ANKHD1 |
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr4_+_170541835 | 0.04 |
ENST00000504131.2 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr10_+_52094298 | 0.04 |
ENST00000595931.1 |
AC069547.1 |
HCG1745369; PRO3073; Uncharacterized protein |
| chr11_-_102576537 | 0.04 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr15_+_75970672 | 0.04 |
ENST00000435356.1 |
AC105020.1 |
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr12_+_54718904 | 0.04 |
ENST00000262061.2 ENST00000549043.1 ENST00000552218.1 ENST00000553231.1 ENST00000552362.1 ENST00000455864.2 ENST00000416254.2 ENST00000549116.1 ENST00000551779.1 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
| chrX_+_69488174 | 0.04 |
ENST00000480877.2 ENST00000307959.8 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr20_+_1875942 | 0.04 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr19_+_10531150 | 0.04 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr11_+_2323349 | 0.04 |
ENST00000381121.3 |
TSPAN32 |
tetraspanin 32 |
| chr1_-_152086556 | 0.03 |
ENST00000368804.1 |
TCHH |
trichohyalin |
| chr3_-_54962100 | 0.03 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr9_-_132404374 | 0.03 |
ENST00000277459.4 ENST00000450050.2 ENST00000277458.4 |
ASB6 |
ankyrin repeat and SOCS box containing 6 |
| chr1_+_35220613 | 0.03 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr6_-_84418860 | 0.03 |
ENST00000521743.1 |
SNAP91 |
synaptosomal-associated protein, 91kDa |
| chr7_-_99573677 | 0.03 |
ENST00000292401.4 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chrX_-_49121165 | 0.03 |
ENST00000376207.4 ENST00000376199.2 |
FOXP3 |
forkhead box P3 |
| chr1_-_13115578 | 0.03 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr17_-_7080227 | 0.03 |
ENST00000574330.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr19_+_56165480 | 0.03 |
ENST00000450554.2 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
| chr3_+_15468862 | 0.02 |
ENST00000396842.2 |
EAF1 |
ELL associated factor 1 |
| chr6_-_84418841 | 0.02 |
ENST00000369694.2 ENST00000195649.6 |
SNAP91 |
synaptosomal-associated protein, 91kDa |
| chr17_+_7531281 | 0.02 |
ENST00000575729.1 ENST00000340624.5 |
SHBG |
sex hormone-binding globulin |
| chr1_-_45805667 | 0.02 |
ENST00000488731.2 ENST00000435155.1 |
MUTYH |
mutY homolog |
| chr12_-_103310987 | 0.02 |
ENST00000307000.2 |
PAH |
phenylalanine hydroxylase |
| chrX_-_53449593 | 0.02 |
ENST00000375340.6 ENST00000322213.4 |
SMC1A |
structural maintenance of chromosomes 1A |
| chr17_+_41150290 | 0.02 |
ENST00000589037.1 ENST00000253788.5 |
RPL27 |
ribosomal protein L27 |
| chr1_+_50569575 | 0.02 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr19_-_4831701 | 0.02 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chrX_+_153448107 | 0.02 |
ENST00000369935.5 |
OPN1MW |
opsin 1 (cone pigments), medium-wave-sensitive |
| chr5_+_139781393 | 0.01 |
ENST00000360839.2 ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
| chr5_+_78365577 | 0.01 |
ENST00000518666.1 ENST00000521567.1 |
BHMT2 |
betaine--homocysteine S-methyltransferase 2 |
| chrX_-_70474499 | 0.01 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr1_-_184943610 | 0.01 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr1_+_247582097 | 0.01 |
ENST00000391827.2 |
NLRP3 |
NLR family, pyrin domain containing 3 |
| chr9_-_22009241 | 0.01 |
ENST00000380142.4 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr2_+_3622932 | 0.01 |
ENST00000406376.1 |
RPS7 |
ribosomal protein S7 |
| chr1_+_20396649 | 0.01 |
ENST00000375108.3 |
PLA2G5 |
phospholipase A2, group V |
| chr18_-_3845321 | 0.01 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 1.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 3.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 5.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.9 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 2.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.5 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.2 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 1.0 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 2.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 | 0.9 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.4 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 3.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.3 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 1.8 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.2 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.5 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 1.0 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0006109 | regulation of carbohydrate metabolic process(GO:0006109) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0031344 | regulation of cell projection organization(GO:0031344) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.5 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |