Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
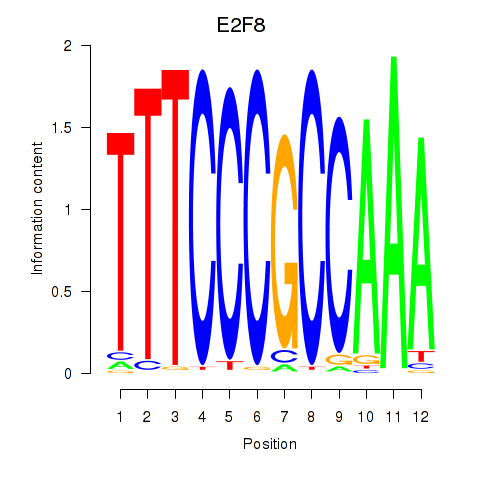
Results for E2F8
Z-value: 1.18
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19263145_19263176 | 0.75 | 8.1e-04 | Click! |
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_135502408 | 7.48 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_+_135502466 | 6.90 |
ENST00000367814.4 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr10_+_70847852 | 3.61 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr6_+_135502501 | 3.29 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chrX_+_52240504 | 3.09 |
ENST00000399805.2 |
XAGE1B |
X antigen family, member 1B |
| chr22_+_19467261 | 2.89 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chrX_-_52736211 | 2.70 |
ENST00000336777.5 ENST00000337502.5 |
SSX2 |
synovial sarcoma, X breakpoint 2 |
| chrX_-_52683950 | 2.53 |
ENST00000298181.5 |
SSX7 |
synovial sarcoma, X breakpoint 7 |
| chr20_+_42295745 | 2.06 |
ENST00000396863.4 ENST00000217026.4 |
MYBL2 |
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr2_-_152146385 | 2.01 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chrX_-_48216101 | 1.94 |
ENST00000298396.2 ENST00000376893.3 |
SSX3 |
synovial sarcoma, X breakpoint 3 |
| chrX_-_153707246 | 1.63 |
ENST00000407062.1 |
LAGE3 |
L antigen family, member 3 |
| chrX_-_153707545 | 1.59 |
ENST00000357360.4 |
LAGE3 |
L antigen family, member 3 |
| chr11_+_125496124 | 1.59 |
ENST00000533778.2 ENST00000534070.1 |
CHEK1 |
checkpoint kinase 1 |
| chr11_+_125495862 | 1.58 |
ENST00000428830.2 ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1 |
checkpoint kinase 1 |
| chr9_+_100263912 | 1.56 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chr1_-_28241024 | 1.49 |
ENST00000313433.7 ENST00000444045.1 |
RPA2 |
replication protein A2, 32kDa |
| chr19_+_50887585 | 1.46 |
ENST00000440232.2 ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1 |
polymerase (DNA directed), delta 1, catalytic subunit |
| chr4_-_174254823 | 1.44 |
ENST00000438704.2 |
HMGB2 |
high mobility group box 2 |
| chr8_-_120868078 | 1.42 |
ENST00000313655.4 |
DSCC1 |
DNA replication and sister chromatid cohesion 1 |
| chr12_-_57146095 | 1.41 |
ENST00000550770.1 ENST00000338193.6 |
PRIM1 |
primase, DNA, polypeptide 1 (49kDa) |
| chr11_-_19263145 | 1.40 |
ENST00000532666.1 ENST00000527884.1 |
E2F8 |
E2F transcription factor 8 |
| chr17_-_46623441 | 1.40 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr16_-_103572 | 1.38 |
ENST00000293860.5 |
POLR3K |
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chrX_-_48271344 | 1.33 |
ENST00000376884.2 ENST00000396928.1 |
SSX4B |
synovial sarcoma, X breakpoint 4B |
| chr22_+_35796108 | 1.28 |
ENST00000382011.5 ENST00000416905.1 |
MCM5 |
minichromosome maintenance complex component 5 |
| chr2_-_136633940 | 1.27 |
ENST00000264156.2 |
MCM6 |
minichromosome maintenance complex component 6 |
| chr6_+_26251835 | 1.26 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr7_+_135242652 | 1.22 |
ENST00000285968.6 ENST00000440390.2 |
NUP205 |
nucleoporin 205kDa |
| chr22_+_35796056 | 1.21 |
ENST00000216122.4 |
MCM5 |
minichromosome maintenance complex component 5 |
| chr14_-_55658252 | 1.18 |
ENST00000395425.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr3_-_25824925 | 1.15 |
ENST00000396649.3 ENST00000428257.1 ENST00000280700.5 |
NGLY1 |
N-glycanase 1 |
| chr8_+_48873479 | 1.13 |
ENST00000262105.2 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr15_+_84115868 | 1.10 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr8_+_48873453 | 1.09 |
ENST00000523944.1 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr12_-_54071181 | 1.02 |
ENST00000338662.5 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr1_-_158656488 | 1.00 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr1_-_159893507 | 0.99 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chr17_-_43128943 | 0.97 |
ENST00000588499.1 ENST00000593094.1 |
DCAKD |
dephospho-CoA kinase domain containing |
| chr3_+_69985792 | 0.96 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr5_-_79950371 | 0.92 |
ENST00000511032.1 ENST00000504396.1 ENST00000505337.1 |
DHFR |
dihydrofolate reductase |
| chr14_-_55658323 | 0.92 |
ENST00000554067.1 ENST00000247191.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr7_+_116593568 | 0.91 |
ENST00000446490.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr7_+_116593433 | 0.86 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr1_+_25598989 | 0.86 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chr15_+_44119159 | 0.82 |
ENST00000263795.6 ENST00000381246.2 ENST00000452115.1 |
WDR76 |
WD repeat domain 76 |
| chr10_+_35416090 | 0.82 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr3_+_127317066 | 0.81 |
ENST00000265056.7 |
MCM2 |
minichromosome maintenance complex component 2 |
| chr12_+_123011776 | 0.80 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr5_+_31532373 | 0.79 |
ENST00000325366.9 ENST00000355907.3 ENST00000507818.2 |
C5orf22 |
chromosome 5 open reading frame 22 |
| chr17_+_76165213 | 0.79 |
ENST00000590201.1 |
SYNGR2 |
synaptogyrin 2 |
| chr1_-_28241226 | 0.77 |
ENST00000373912.3 ENST00000373909.3 |
RPA2 |
replication protein A2, 32kDa |
| chr11_+_65029233 | 0.76 |
ENST00000265465.3 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr10_-_43892279 | 0.74 |
ENST00000443950.2 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr14_-_23388338 | 0.73 |
ENST00000555209.1 ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23 |
RNA binding motif protein 23 |
| chr17_+_56769924 | 0.72 |
ENST00000461271.1 ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C |
RAD51 paralog C |
| chr1_+_33283043 | 0.72 |
ENST00000373476.1 ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP |
S100P binding protein |
| chr15_+_84116106 | 0.71 |
ENST00000535412.1 ENST00000324537.5 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr8_+_128748308 | 0.70 |
ENST00000377970.2 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
| chr9_+_131445928 | 0.69 |
ENST00000372692.4 |
SET |
SET nuclear oncogene |
| chr15_+_64428529 | 0.68 |
ENST00000560861.1 |
SNX1 |
sorting nexin 1 |
| chr22_+_20105012 | 0.67 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr15_+_66797627 | 0.67 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr8_+_128748466 | 0.67 |
ENST00000524013.1 ENST00000520751.1 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
| chr4_-_1713977 | 0.66 |
ENST00000318386.4 |
SLBP |
stem-loop binding protein |
| chr14_-_100841670 | 0.64 |
ENST00000557297.1 ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr4_+_128702969 | 0.64 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chrX_+_131157290 | 0.63 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr1_-_26232522 | 0.63 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr2_-_215674374 | 0.62 |
ENST00000449967.2 ENST00000421162.1 ENST00000260947.4 |
BARD1 |
BRCA1 associated RING domain 1 |
| chr4_-_106629796 | 0.62 |
ENST00000416543.1 ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12 |
integrator complex subunit 12 |
| chrX_+_24072833 | 0.61 |
ENST00000253039.4 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr7_+_26331541 | 0.61 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr17_-_30668887 | 0.61 |
ENST00000581747.1 ENST00000583334.1 ENST00000580558.1 |
C17orf75 |
chromosome 17 open reading frame 75 |
| chr13_-_113862401 | 0.60 |
ENST00000375459.1 |
PCID2 |
PCI domain containing 2 |
| chrX_+_41192595 | 0.60 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr6_+_10748019 | 0.60 |
ENST00000543878.1 ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L TMEM14B |
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr4_-_1714037 | 0.59 |
ENST00000488267.1 ENST00000429429.2 ENST00000480936.1 |
SLBP |
stem-loop binding protein |
| chr6_-_32157947 | 0.59 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr10_+_35416223 | 0.59 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chrX_+_131157322 | 0.59 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_-_129299847 | 0.59 |
ENST00000319908.3 ENST00000287295.3 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr2_+_10262857 | 0.58 |
ENST00000304567.5 |
RRM2 |
ribonucleotide reductase M2 |
| chr1_+_70876926 | 0.58 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr12_-_49245936 | 0.58 |
ENST00000308025.3 |
DDX23 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr10_-_104178857 | 0.58 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr1_+_25598872 | 0.57 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr17_+_5390220 | 0.57 |
ENST00000381165.3 |
MIS12 |
MIS12 kinetochore complex component |
| chr16_+_103816 | 0.56 |
ENST00000383018.3 ENST00000417493.1 |
SNRNP25 |
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr12_+_107168418 | 0.56 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr6_+_10747986 | 0.56 |
ENST00000379542.5 |
TMEM14B |
transmembrane protein 14B |
| chr10_+_104178946 | 0.55 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr3_+_14989076 | 0.55 |
ENST00000413118.1 ENST00000425241.1 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_25599018 | 0.55 |
ENST00000417538.2 ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD |
Rh blood group, D antigen |
| chr12_+_111843749 | 0.54 |
ENST00000341259.2 |
SH2B3 |
SH2B adaptor protein 3 |
| chr6_+_127588020 | 0.53 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr6_+_43603552 | 0.52 |
ENST00000372171.4 |
MAD2L1BP |
MAD2L1 binding protein |
| chr6_-_111804905 | 0.51 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr1_+_97187318 | 0.51 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr18_-_47018897 | 0.51 |
ENST00000418495.1 |
RPL17 |
ribosomal protein L17 |
| chr4_+_25314388 | 0.50 |
ENST00000302874.4 |
ZCCHC4 |
zinc finger, CCHC domain containing 4 |
| chr18_-_47018869 | 0.49 |
ENST00000583036.1 ENST00000580261.1 |
RPL17 |
ribosomal protein L17 |
| chr1_+_70876891 | 0.49 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr15_+_40731920 | 0.49 |
ENST00000561234.1 |
BAHD1 |
bromo adjacent homology domain containing 1 |
| chr1_-_6295975 | 0.48 |
ENST00000343813.5 ENST00000362035.3 |
ICMT |
isoprenylcysteine carboxyl methyltransferase |
| chr6_-_33290580 | 0.48 |
ENST00000446511.1 ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX |
death-domain associated protein |
| chr1_+_154975110 | 0.48 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr17_-_29151794 | 0.48 |
ENST00000324238.6 |
CRLF3 |
cytokine receptor-like factor 3 |
| chr4_+_57845043 | 0.47 |
ENST00000433463.1 ENST00000314595.5 |
POLR2B |
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr15_-_64673630 | 0.45 |
ENST00000558008.1 ENST00000559519.1 ENST00000380258.2 |
KIAA0101 |
KIAA0101 |
| chr15_-_89456630 | 0.44 |
ENST00000268150.8 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr8_+_29953163 | 0.44 |
ENST00000518192.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr19_-_3500635 | 0.44 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr1_-_43638168 | 0.43 |
ENST00000431635.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr3_+_184098065 | 0.43 |
ENST00000348986.3 |
CHRD |
chordin |
| chr4_+_108910870 | 0.43 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr6_-_26659913 | 0.42 |
ENST00000480036.1 ENST00000415922.2 |
ZNF322 |
zinc finger protein 322 |
| chr22_-_20104700 | 0.41 |
ENST00000439169.2 ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr10_-_27443155 | 0.41 |
ENST00000427324.1 ENST00000326799.3 |
YME1L1 |
YME1-like 1 ATPase |
| chr22_+_40742497 | 0.41 |
ENST00000216194.7 |
ADSL |
adenylosuccinate lyase |
| chr19_+_2270283 | 0.41 |
ENST00000588673.2 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
| chr12_+_123459127 | 0.41 |
ENST00000397389.2 ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2 |
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr2_+_132233664 | 0.41 |
ENST00000321253.6 |
TUBA3D |
tubulin, alpha 3d |
| chr4_+_57845024 | 0.40 |
ENST00000431623.2 ENST00000441246.2 |
POLR2B |
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr11_+_134201911 | 0.39 |
ENST00000389881.3 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr3_-_194393206 | 0.37 |
ENST00000265245.5 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr6_-_57086200 | 0.37 |
ENST00000468148.1 |
RAB23 |
RAB23, member RAS oncogene family |
| chr3_+_44803209 | 0.36 |
ENST00000326047.4 |
KIF15 |
kinesin family member 15 |
| chr17_-_2415169 | 0.36 |
ENST00000263092.6 ENST00000538844.1 ENST00000576976.1 |
METTL16 |
methyltransferase like 16 |
| chr20_-_2644832 | 0.36 |
ENST00000380851.5 ENST00000380843.4 |
IDH3B |
isocitrate dehydrogenase 3 (NAD+) beta |
| chr15_-_89456593 | 0.36 |
ENST00000558029.1 ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr17_+_5389605 | 0.36 |
ENST00000576988.1 ENST00000576570.1 ENST00000573759.1 |
MIS12 |
MIS12 kinetochore complex component |
| chr19_+_4402659 | 0.35 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr22_+_40742512 | 0.33 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr13_-_19755975 | 0.33 |
ENST00000400113.3 |
TUBA3C |
tubulin, alpha 3c |
| chr6_+_70576457 | 0.32 |
ENST00000322773.4 |
COL19A1 |
collagen, type XIX, alpha 1 |
| chr11_+_118992269 | 0.31 |
ENST00000350777.2 ENST00000529988.1 ENST00000527410.1 |
HINFP |
histone H4 transcription factor |
| chr7_+_119913688 | 0.31 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr2_-_148778323 | 0.30 |
ENST00000440042.1 ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4 |
origin recognition complex, subunit 4 |
| chr4_-_140223670 | 0.30 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_-_130956006 | 0.29 |
ENST00000312988.7 |
TUBA3E |
tubulin, alpha 3e |
| chr10_+_115312766 | 0.29 |
ENST00000351270.3 |
HABP2 |
hyaluronan binding protein 2 |
| chr15_-_34635314 | 0.29 |
ENST00000557912.1 ENST00000328848.4 |
NOP10 |
NOP10 ribonucleoprotein |
| chr17_+_61904766 | 0.29 |
ENST00000581842.1 ENST00000582130.1 ENST00000584320.1 ENST00000585123.1 ENST00000580864.1 |
PSMC5 |
proteasome (prosome, macropain) 26S subunit, ATPase, 5 |
| chr3_+_197677047 | 0.29 |
ENST00000448864.1 |
RPL35A |
ribosomal protein L35a |
| chr8_+_29952914 | 0.29 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr14_+_51706886 | 0.29 |
ENST00000457354.2 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
| chr2_-_148778258 | 0.28 |
ENST00000392857.5 ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4 |
origin recognition complex, subunit 4 |
| chr15_-_64673665 | 0.28 |
ENST00000300035.4 |
KIAA0101 |
KIAA0101 |
| chr4_-_140223614 | 0.27 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr10_-_27443294 | 0.27 |
ENST00000396296.3 ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1 |
YME1-like 1 ATPase |
| chr3_+_14989186 | 0.27 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr3_-_125313934 | 0.27 |
ENST00000296220.5 |
OSBPL11 |
oxysterol binding protein-like 11 |
| chr3_+_133293278 | 0.26 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr9_-_136039282 | 0.26 |
ENST00000372036.3 ENST00000372038.3 ENST00000540636.1 ENST00000372043.3 ENST00000542690.1 |
GBGT1 RALGDS |
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 ral guanine nucleotide dissociation stimulator |
| chr16_-_66864806 | 0.26 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr12_+_56521840 | 0.26 |
ENST00000394048.5 |
ESYT1 |
extended synaptotagmin-like protein 1 |
| chr11_+_71938925 | 0.26 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr15_-_25684110 | 0.25 |
ENST00000232165.3 |
UBE3A |
ubiquitin protein ligase E3A |
| chr2_-_128051670 | 0.25 |
ENST00000493187.2 |
ERCC3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr6_+_26204825 | 0.25 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr4_-_129209944 | 0.25 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr4_-_4543700 | 0.24 |
ENST00000505286.1 ENST00000306200.2 |
STX18 |
syntaxin 18 |
| chr2_-_58468437 | 0.24 |
ENST00000403676.1 ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL |
Fanconi anemia, complementation group L |
| chr3_+_19988566 | 0.24 |
ENST00000273047.4 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr17_-_15165854 | 0.24 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr19_-_46146946 | 0.24 |
ENST00000536630.1 |
EML2 |
echinoderm microtubule associated protein like 2 |
| chr2_+_63816295 | 0.23 |
ENST00000539945.1 ENST00000544381.1 |
MDH1 |
malate dehydrogenase 1, NAD (soluble) |
| chr16_+_56995762 | 0.23 |
ENST00000200676.3 ENST00000379780.2 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr2_-_128051708 | 0.23 |
ENST00000285398.2 |
ERCC3 |
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chr12_-_16761007 | 0.21 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_205719295 | 0.21 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr3_+_70048881 | 0.21 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr7_+_116593292 | 0.21 |
ENST00000393446.2 ENST00000265437.5 ENST00000393451.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr13_-_41345277 | 0.20 |
ENST00000323563.6 |
MRPS31 |
mitochondrial ribosomal protein S31 |
| chr19_+_49403562 | 0.20 |
ENST00000407032.1 ENST00000452087.1 ENST00000411700.1 |
NUCB1 |
nucleobindin 1 |
| chr11_-_19262486 | 0.19 |
ENST00000250024.4 |
E2F8 |
E2F transcription factor 8 |
| chr7_+_128379449 | 0.19 |
ENST00000479257.1 |
CALU |
calumenin |
| chr6_-_2962331 | 0.18 |
ENST00000380524.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr7_-_140714739 | 0.18 |
ENST00000467334.1 ENST00000324787.5 |
MRPS33 |
mitochondrial ribosomal protein S33 |
| chr5_-_58882219 | 0.18 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr17_-_59940830 | 0.18 |
ENST00000259008.2 |
BRIP1 |
BRCA1 interacting protein C-terminal helicase 1 |
| chr1_-_43637915 | 0.18 |
ENST00000236051.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr4_-_176733897 | 0.18 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr19_-_19249255 | 0.17 |
ENST00000587583.2 ENST00000450333.2 ENST00000587096.1 ENST00000162044.9 ENST00000592369.1 ENST00000587915.1 |
TMEM161A |
transmembrane protein 161A |
| chr17_+_16120512 | 0.17 |
ENST00000581006.1 ENST00000584797.1 ENST00000498772.2 ENST00000225609.5 ENST00000395844.4 ENST00000477745.1 |
PIGL |
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr12_+_3069037 | 0.17 |
ENST00000397122.2 |
TEAD4 |
TEA domain family member 4 |
| chr1_+_150245099 | 0.17 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr22_-_42086477 | 0.17 |
ENST00000402458.1 |
NHP2L1 |
NHP2 non-histone chromosome protein 2-like 1 (S. cerevisiae) |
| chrX_-_119695279 | 0.17 |
ENST00000336592.6 |
CUL4B |
cullin 4B |
| chr22_-_36877629 | 0.17 |
ENST00000416967.1 |
TXN2 |
thioredoxin 2 |
| chr17_-_40897043 | 0.16 |
ENST00000428826.2 ENST00000592492.1 ENST00000585893.1 ENST00000593214.1 ENST00000590078.1 ENST00000586382.1 ENST00000415827.2 ENST00000592743.1 ENST00000586089.1 ENST00000435174.1 |
EZH1 |
enhancer of zeste homolog 1 (Drosophila) |
| chr1_-_6453399 | 0.16 |
ENST00000608083.1 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr11_-_89224508 | 0.16 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr14_+_60975644 | 0.15 |
ENST00000327720.5 |
SIX6 |
SIX homeobox 6 |
| chr3_-_137893721 | 0.15 |
ENST00000505015.2 ENST00000260803.4 |
DBR1 |
debranching RNA lariats 1 |
| chr18_-_47017956 | 0.15 |
ENST00000584895.1 ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32 RPL17 |
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr4_-_84406218 | 0.15 |
ENST00000515303.1 |
FAM175A |
family with sequence similarity 175, member A |
| chr7_+_75932863 | 0.14 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr1_-_89458287 | 0.14 |
ENST00000370485.2 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 5.9 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.2 | 3.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 19.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 3.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.4 | 6.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 1.4 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.2 | 1.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 1.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.2 | 1.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.9 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.6 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 1.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 4.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.5 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 3.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 2.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 2.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 8.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 17.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 5.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 2.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.6 | 3.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.5 | 6.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 2.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 1.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 1.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 0.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 3.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 2.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.6 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 1.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.8 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 16.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.6 | GO:0008180 | striated muscle thin filament(GO:0005865) COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.7 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 1.2 | 3.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.0 | 2.9 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.7 | 2.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.5 | 3.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.5 | 1.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.4 | 4.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 2.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.3 | 1.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 1.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 1.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.3 | 1.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 0.9 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.2 | 1.4 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.2 | 0.6 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 3.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 1.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.4 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.5 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.1 | 0.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 3.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.6 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.1 | 0.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.6 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.2 | GO:0071812 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 2.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 1.4 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.4 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 3.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.1 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 1.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.0 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.1 | GO:0002666 | positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.2 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 1.0 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.0 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |