Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for EBF3
Z-value: 0.91
Transcription factors associated with EBF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EBF3
|
ENSG00000108001.9 | EBF3 |
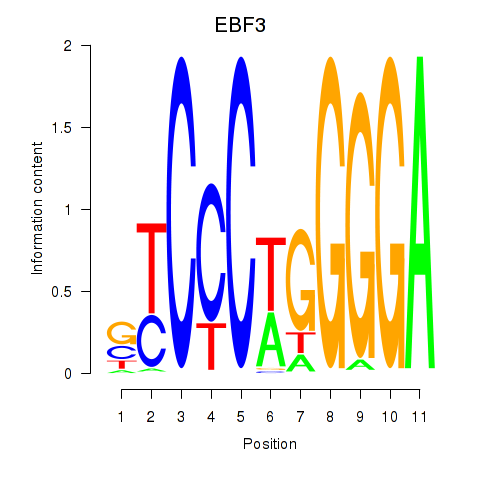
Activity profile of EBF3 motif
Sorted Z-values of EBF3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EBF3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_101193246 | 3.61 |
ENST00000331224.6 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr14_+_101193164 | 3.57 |
ENST00000341267.4 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr11_+_116700614 | 2.11 |
ENST00000375345.1 |
APOC3 |
apolipoprotein C-III |
| chr11_+_116700600 | 2.11 |
ENST00000227667.3 |
APOC3 |
apolipoprotein C-III |
| chr17_-_7082861 | 1.84 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr11_+_22689648 | 1.50 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr17_-_7082668 | 1.50 |
ENST00000573083.1 ENST00000574388.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr17_-_56494908 | 1.47 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494882 | 1.46 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr2_-_238499303 | 1.45 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr16_+_72088376 | 1.41 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr17_-_56494713 | 1.38 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr17_+_41006095 | 1.37 |
ENST00000591562.1 ENST00000588033.1 |
AOC3 |
amine oxidase, copper containing 3 |
| chrX_+_105937068 | 1.34 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_-_100240328 | 1.33 |
ENST00000462107.1 |
TFR2 |
transferrin receptor 2 |
| chr1_-_21995794 | 1.33 |
ENST00000542643.2 ENST00000374765.4 ENST00000317967.7 |
RAP1GAP |
RAP1 GTPase activating protein |
| chrX_+_192989 | 1.25 |
ENST00000399012.1 ENST00000430923.2 |
PLCXD1 |
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr16_-_68269971 | 1.19 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr16_-_30798492 | 1.18 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chr1_-_182361327 | 1.08 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr1_-_182360918 | 1.06 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr4_+_1795012 | 1.03 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr5_-_175964366 | 1.01 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr8_-_145641864 | 0.91 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr4_+_4861385 | 0.90 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr8_+_21912328 | 0.89 |
ENST00000432128.1 ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN |
dematin actin binding protein |
| chrX_+_38420623 | 0.86 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr17_-_79881408 | 0.82 |
ENST00000392366.3 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr2_+_236402669 | 0.81 |
ENST00000409457.1 ENST00000336665.5 ENST00000304032.8 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr3_+_193853927 | 0.81 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr1_+_2004901 | 0.81 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr2_+_128177458 | 0.80 |
ENST00000409048.1 ENST00000422777.3 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr14_+_24590560 | 0.80 |
ENST00000558325.1 |
RP11-468E2.6 |
RP11-468E2.6 |
| chrX_+_46940254 | 0.75 |
ENST00000336169.3 |
RGN |
regucalcin |
| chr18_-_28681950 | 0.73 |
ENST00000251081.6 |
DSC2 |
desmocollin 2 |
| chr1_+_109792641 | 0.72 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr2_-_238499131 | 0.71 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr22_+_38201114 | 0.70 |
ENST00000340857.2 |
H1F0 |
H1 histone family, member 0 |
| chr12_-_54694758 | 0.69 |
ENST00000553070.1 |
NFE2 |
nuclear factor, erythroid 2 |
| chrX_+_38420783 | 0.69 |
ENST00000422612.2 ENST00000286824.6 ENST00000545599.1 |
TSPAN7 |
tetraspanin 7 |
| chr2_-_238499725 | 0.69 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr7_-_71912046 | 0.69 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr10_+_114133773 | 0.67 |
ENST00000354655.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr22_-_30960876 | 0.67 |
ENST00000401975.1 ENST00000428682.1 ENST00000423299.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr1_+_2005425 | 0.65 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chrX_+_152990302 | 0.64 |
ENST00000218104.3 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr2_-_188378368 | 0.64 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_+_100897130 | 0.64 |
ENST00000551379.1 ENST00000188403.7 ENST00000551184.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr11_-_117699413 | 0.64 |
ENST00000528014.1 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr7_-_158380465 | 0.62 |
ENST00000389413.3 ENST00000409483.1 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr11_-_72432950 | 0.61 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr7_-_158380371 | 0.60 |
ENST00000389418.4 ENST00000389416.4 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr3_-_149293990 | 0.60 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr11_-_117698787 | 0.60 |
ENST00000260287.2 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr7_-_148823387 | 0.60 |
ENST00000483014.1 ENST00000378061.2 |
ZNF425 |
zinc finger protein 425 |
| chr3_+_52813932 | 0.59 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr12_-_54694807 | 0.59 |
ENST00000435572.2 |
NFE2 |
nuclear factor, erythroid 2 |
| chr11_-_117698765 | 0.59 |
ENST00000532119.1 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chrX_+_69509927 | 0.59 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr2_-_238499337 | 0.58 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr11_-_65381643 | 0.56 |
ENST00000309100.3 ENST00000529839.1 ENST00000526293.1 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
| chr1_+_46049706 | 0.56 |
ENST00000527470.1 ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
| chr11_+_1860832 | 0.54 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr15_+_66994561 | 0.54 |
ENST00000288840.5 |
SMAD6 |
SMAD family member 6 |
| chr2_-_64371546 | 0.54 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr1_+_174769006 | 0.53 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr6_+_42984723 | 0.52 |
ENST00000332245.8 |
KLHDC3 |
kelch domain containing 3 |
| chr17_-_34890709 | 0.51 |
ENST00000544606.1 |
MYO19 |
myosin XIX |
| chrX_+_48644962 | 0.51 |
ENST00000376670.3 ENST00000376665.3 |
GATA1 |
GATA binding protein 1 (globin transcription factor 1) |
| chr1_+_236687881 | 0.51 |
ENST00000526634.1 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr10_+_114710516 | 0.50 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_-_152989798 | 0.49 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr1_+_28696111 | 0.49 |
ENST00000373839.3 |
PHACTR4 |
phosphatase and actin regulator 4 |
| chr2_-_27632390 | 0.49 |
ENST00000350803.4 ENST00000344034.4 |
PPM1G |
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr2_-_174830430 | 0.48 |
ENST00000310015.6 ENST00000455789.2 |
SP3 |
Sp3 transcription factor |
| chr22_+_39898325 | 0.47 |
ENST00000325301.2 ENST00000404569.1 |
MIEF1 |
mitochondrial elongation factor 1 |
| chr2_-_128785688 | 0.47 |
ENST00000259234.6 |
SAP130 |
Sin3A-associated protein, 130kDa |
| chr12_-_71182695 | 0.47 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr6_+_43737939 | 0.47 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr1_-_117210290 | 0.46 |
ENST00000369483.1 ENST00000369486.3 |
IGSF3 |
immunoglobulin superfamily, member 3 |
| chr17_-_42580738 | 0.46 |
ENST00000585614.1 ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8 |
G patch domain containing 8 |
| chr7_-_100881109 | 0.46 |
ENST00000308344.5 |
CLDN15 |
claudin 15 |
| chr7_-_98467629 | 0.46 |
ENST00000339375.4 |
TMEM130 |
transmembrane protein 130 |
| chr1_+_26869597 | 0.46 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr11_+_1860682 | 0.46 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr17_-_5026397 | 0.45 |
ENST00000250076.3 |
ZNF232 |
zinc finger protein 232 |
| chr1_+_14925173 | 0.45 |
ENST00000376030.2 ENST00000503743.1 ENST00000422387.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr6_+_43738444 | 0.44 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr7_-_98467543 | 0.44 |
ENST00000345589.4 |
TMEM130 |
transmembrane protein 130 |
| chr6_+_7107999 | 0.44 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr3_-_128212016 | 0.44 |
ENST00000498200.1 ENST00000341105.2 |
GATA2 |
GATA binding protein 2 |
| chr4_+_37892682 | 0.43 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr1_+_110453109 | 0.43 |
ENST00000525659.1 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chrX_-_33229636 | 0.43 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr1_+_150229554 | 0.42 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr16_+_23569021 | 0.42 |
ENST00000567212.1 ENST00000567264.1 |
UBFD1 |
ubiquitin family domain containing 1 |
| chr17_-_80023401 | 0.41 |
ENST00000354321.7 ENST00000306796.5 |
DUS1L |
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr10_+_114709999 | 0.41 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr3_-_185826855 | 0.40 |
ENST00000306376.5 |
ETV5 |
ets variant 5 |
| chr5_+_154393260 | 0.40 |
ENST00000435029.4 |
KIF4B |
kinesin family member 4B |
| chrX_-_74376108 | 0.40 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr16_+_28505955 | 0.40 |
ENST00000564831.1 ENST00000328423.5 ENST00000431282.1 |
APOBR |
apolipoprotein B receptor |
| chr16_+_72142195 | 0.39 |
ENST00000563819.1 ENST00000567142.2 |
DHX38 |
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr1_+_11751748 | 0.38 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr11_-_72433346 | 0.38 |
ENST00000334211.8 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr14_+_23790655 | 0.38 |
ENST00000397276.2 |
PABPN1 |
poly(A) binding protein, nuclear 1 |
| chr14_+_105953246 | 0.37 |
ENST00000392531.3 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr19_+_33865218 | 0.37 |
ENST00000585933.2 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr17_+_73452545 | 0.37 |
ENST00000314256.7 |
KIAA0195 |
KIAA0195 |
| chr7_-_1015198 | 0.37 |
ENST00000344111.3 |
COX19 |
cytochrome c oxidase assembly homolog 19 (S. cerevisiae) |
| chr19_-_55881741 | 0.36 |
ENST00000264563.2 ENST00000590625.1 ENST00000585513.1 |
IL11 |
interleukin 11 |
| chr15_+_90544532 | 0.36 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chr20_-_30795511 | 0.36 |
ENST00000246229.4 |
PLAGL2 |
pleiomorphic adenoma gene-like 2 |
| chr14_+_105953204 | 0.35 |
ENST00000409393.2 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr9_+_134103496 | 0.35 |
ENST00000498010.1 ENST00000476004.1 ENST00000528406.1 |
NUP214 |
nucleoporin 214kDa |
| chr16_+_30662184 | 0.35 |
ENST00000300835.4 |
PRR14 |
proline rich 14 |
| chr18_+_10454594 | 0.35 |
ENST00000355285.5 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
| chr2_+_169312350 | 0.34 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr8_-_80680078 | 0.34 |
ENST00000337919.5 ENST00000354724.3 |
HEY1 |
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr17_+_43972010 | 0.34 |
ENST00000334239.8 ENST00000446361.3 |
MAPT |
microtubule-associated protein tau |
| chr19_-_59010565 | 0.34 |
ENST00000594786.1 |
SLC27A5 |
solute carrier family 27 (fatty acid transporter), member 5 |
| chr7_+_29874341 | 0.34 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chr10_+_88428370 | 0.33 |
ENST00000372066.3 ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3 |
LIM domain binding 3 |
| chr13_+_113622810 | 0.33 |
ENST00000397030.1 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr13_+_113622757 | 0.33 |
ENST00000375604.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr14_+_75536335 | 0.33 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr1_-_151431909 | 0.33 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr12_+_7282795 | 0.33 |
ENST00000266546.6 |
CLSTN3 |
calsyntenin 3 |
| chr12_+_1100370 | 0.32 |
ENST00000543086.3 ENST00000546231.2 ENST00000397203.2 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_23540826 | 0.32 |
ENST00000357481.2 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
| chr1_+_3388181 | 0.32 |
ENST00000418137.1 ENST00000413250.2 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr10_-_73848764 | 0.32 |
ENST00000317376.4 ENST00000412663.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr19_-_7293942 | 0.31 |
ENST00000341500.5 ENST00000302850.5 |
INSR |
insulin receptor |
| chr14_+_105952648 | 0.31 |
ENST00000330233.7 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr1_-_151431647 | 0.30 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr9_+_136325089 | 0.30 |
ENST00000291722.7 ENST00000316948.4 ENST00000540581.1 |
CACFD1 |
calcium channel flower domain containing 1 |
| chr8_+_28748099 | 0.30 |
ENST00000519047.1 |
HMBOX1 |
homeobox containing 1 |
| chr7_+_107110488 | 0.30 |
ENST00000304402.4 |
GPR22 |
G protein-coupled receptor 22 |
| chr2_+_27070964 | 0.29 |
ENST00000288699.6 |
DPYSL5 |
dihydropyrimidinase-like 5 |
| chr16_-_68482440 | 0.29 |
ENST00000219334.5 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr9_+_135854091 | 0.29 |
ENST00000450530.1 ENST00000534944.1 |
GFI1B |
growth factor independent 1B transcription repressor |
| chr14_+_75536280 | 0.29 |
ENST00000238686.8 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
| chr2_-_219134822 | 0.29 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr10_+_102106829 | 0.28 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr16_+_30662360 | 0.28 |
ENST00000542965.2 |
PRR14 |
proline rich 14 |
| chr11_+_64879317 | 0.28 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chrX_-_48827976 | 0.28 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr3_+_5020801 | 0.28 |
ENST00000256495.3 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
| chr17_-_37382105 | 0.28 |
ENST00000333461.5 |
STAC2 |
SH3 and cysteine rich domain 2 |
| chr10_+_17851362 | 0.28 |
ENST00000331429.2 ENST00000457317.1 |
MRC1L1 |
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr20_-_35492048 | 0.27 |
ENST00000237536.4 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
| chr11_+_63655987 | 0.27 |
ENST00000509502.2 ENST00000512060.1 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr5_-_58295712 | 0.27 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr1_-_1051455 | 0.27 |
ENST00000379339.1 ENST00000480643.1 ENST00000434641.1 ENST00000421241.2 |
C1orf159 |
chromosome 1 open reading frame 159 |
| chr6_+_161412759 | 0.27 |
ENST00000366919.2 ENST00000392142.4 ENST00000366920.2 ENST00000348824.7 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
| chr10_+_104678032 | 0.27 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr8_-_19459993 | 0.26 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_+_28747884 | 0.26 |
ENST00000287701.10 ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1 |
homeobox containing 1 |
| chr19_+_984313 | 0.26 |
ENST00000251289.5 ENST00000587001.2 ENST00000607440.1 |
WDR18 |
WD repeat domain 18 |
| chr17_-_34890732 | 0.26 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr17_+_38465441 | 0.26 |
ENST00000577646.1 ENST00000254066.5 |
RARA |
retinoic acid receptor, alpha |
| chr12_+_112279782 | 0.26 |
ENST00000550735.2 |
MAPKAPK5 |
mitogen-activated protein kinase-activated protein kinase 5 |
| chr22_-_43411106 | 0.25 |
ENST00000453643.1 ENST00000263246.3 ENST00000337959.4 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_185826286 | 0.25 |
ENST00000537818.1 ENST00000422039.1 ENST00000434744.1 |
ETV5 |
ets variant 5 |
| chr9_+_112403088 | 0.25 |
ENST00000448454.2 |
PALM2 |
paralemmin 2 |
| chr16_-_30381580 | 0.25 |
ENST00000409939.3 |
TBC1D10B |
TBC1 domain family, member 10B |
| chr8_+_145582231 | 0.24 |
ENST00000526338.1 ENST00000402965.1 ENST00000534725.1 ENST00000532887.1 ENST00000329994.2 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr17_+_43971643 | 0.24 |
ENST00000344290.5 ENST00000262410.5 ENST00000351559.5 ENST00000340799.5 ENST00000535772.1 ENST00000347967.5 |
MAPT |
microtubule-associated protein tau |
| chr10_+_104678102 | 0.24 |
ENST00000433628.2 |
CNNM2 |
cyclin M2 |
| chr17_-_34890759 | 0.23 |
ENST00000431794.3 |
MYO19 |
myosin XIX |
| chr8_+_145582217 | 0.23 |
ENST00000530047.1 ENST00000527078.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr17_-_42296855 | 0.23 |
ENST00000436088.1 |
UBTF |
upstream binding transcription factor, RNA polymerase I |
| chr14_-_22005018 | 0.23 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr1_+_110453514 | 0.22 |
ENST00000369802.3 ENST00000420111.2 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr14_-_24553834 | 0.22 |
ENST00000397002.2 |
NRL |
neural retina leucine zipper |
| chr16_-_28506840 | 0.22 |
ENST00000569430.1 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
| chr10_-_102090243 | 0.22 |
ENST00000338519.3 ENST00000353274.3 ENST00000318222.3 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr4_+_39408470 | 0.22 |
ENST00000257408.4 |
KLB |
klotho beta |
| chr4_-_140223614 | 0.22 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_+_37407212 | 0.21 |
ENST00000427117.1 ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568 |
zinc finger protein 568 |
| chr7_-_137686791 | 0.21 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr16_-_29478016 | 0.21 |
ENST00000549858.1 ENST00000551411.1 |
RP11-345J4.3 |
Uncharacterized protein |
| chr1_-_41950342 | 0.21 |
ENST00000372587.4 |
EDN2 |
endothelin 2 |
| chrX_+_9217932 | 0.21 |
ENST00000432442.1 |
GS1-519E5.1 |
GS1-519E5.1 |
| chr11_-_46142948 | 0.21 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr17_+_73472575 | 0.21 |
ENST00000375248.5 |
KIAA0195 |
KIAA0195 |
| chr8_+_136470270 | 0.21 |
ENST00000524199.1 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
| chr1_+_110198714 | 0.21 |
ENST00000336075.5 ENST00000326729.5 |
GSTM4 |
glutathione S-transferase mu 4 |
| chr21_-_45682099 | 0.20 |
ENST00000270172.3 ENST00000418993.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr19_+_49403562 | 0.20 |
ENST00000407032.1 ENST00000452087.1 ENST00000411700.1 |
NUCB1 |
nucleobindin 1 |
| chr10_+_111985713 | 0.20 |
ENST00000239007.7 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr14_+_23791159 | 0.20 |
ENST00000557702.1 |
PABPN1 |
poly(A) binding protein, nuclear 1 |
| chr14_-_75536182 | 0.20 |
ENST00000555463.1 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr7_+_65338230 | 0.19 |
ENST00000360768.3 |
VKORC1L1 |
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr8_+_145582633 | 0.19 |
ENST00000540505.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr19_-_40331345 | 0.19 |
ENST00000597224.1 |
FBL |
fibrillarin |
| chr6_+_144904334 | 0.19 |
ENST00000367526.4 |
UTRN |
utrophin |
| chr7_-_44180673 | 0.19 |
ENST00000457314.1 ENST00000447951.1 ENST00000431007.1 |
MYL7 |
myosin, light chain 7, regulatory |
| chr16_+_69958887 | 0.18 |
ENST00000568684.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr17_-_74023474 | 0.18 |
ENST00000301607.3 |
EVPL |
envoplakin |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 1.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 2.1 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.2 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 4.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.5 | 2.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.5 | 1.4 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.4 | 3.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.3 | 1.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 1.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.6 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 0.9 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 4.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.0 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 2.0 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.5 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.6 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 3.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 4.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 4.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0060620 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.1 | 4.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.7 | 3.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.5 | 2.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 1.4 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 1.3 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.3 | 0.9 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.3 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.3 | 0.9 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.3 | 0.8 | GO:2000977 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.3 | 0.8 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.3 | 0.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.2 | 1.0 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 | 1.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 7.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.7 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.2 | 1.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 0.6 | GO:0070858 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) negative regulation of bile acid biosynthetic process(GO:0070858) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 1.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.9 | GO:1904141 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.9 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 1.3 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.7 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 1.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 1.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.5 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 0.5 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.1 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:1900175 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.3 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:0060585 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 1.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 3.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 1.5 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 1.2 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.9 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 1.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.4 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.4 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.4 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.7 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 1.4 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.1 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 1.3 | GO:0016032 | viral process(GO:0016032) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.3 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 1.0 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |