Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
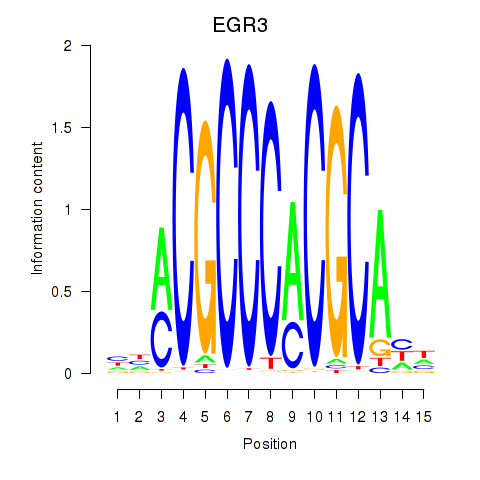
Results for EGR3_EGR2
Z-value: 0.86
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | EGR3 |
|
EGR2
|
ENSG00000122877.9 | EGR2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | -0.66 | 5.5e-03 | Click! |
| EGR3 | hg19_v2_chr8_-_22550815_22550844 | -0.53 | 3.4e-02 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_227506158 | 1.61 |
ENST00000366769.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr1_-_227505826 | 1.40 |
ENST00000334218.5 ENST00000366766.2 ENST00000366764.2 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr15_+_84116106 | 1.31 |
ENST00000535412.1 ENST00000324537.5 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr15_+_84115868 | 1.23 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr1_-_85156216 | 1.20 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr6_-_31697255 | 1.19 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_106685079 | 1.17 |
ENST00000265717.4 |
PRKAR2B |
protein kinase, cAMP-dependent, regulatory, type II, beta |
| chr9_-_33264557 | 0.99 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr10_+_94451574 | 0.97 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr1_-_85156090 | 0.93 |
ENST00000605755.1 ENST00000437941.2 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr6_-_31697563 | 0.93 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr1_+_65886244 | 0.92 |
ENST00000344610.8 |
LEPR |
leptin receptor |
| chrX_-_154033661 | 0.91 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr11_+_124609823 | 0.91 |
ENST00000412681.2 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_114310237 | 0.89 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chrX_-_154033793 | 0.88 |
ENST00000369534.3 ENST00000413259.3 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr11_+_124609742 | 0.84 |
ENST00000284292.6 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_114310164 | 0.83 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr20_-_23066953 | 0.83 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr5_+_34656569 | 0.82 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr9_-_33264676 | 0.81 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr11_+_66624527 | 0.80 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_-_54693401 | 0.80 |
ENST00000338624.6 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chr4_-_90758227 | 0.75 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr20_+_44035200 | 0.75 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_-_69445968 | 0.75 |
ENST00000438964.2 |
ACTN1 |
actinin, alpha 1 |
| chr11_+_114310102 | 0.74 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr14_-_69446034 | 0.73 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr5_+_34656331 | 0.73 |
ENST00000265109.3 |
RAI14 |
retinoic acid induced 14 |
| chr6_-_112575912 | 0.73 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr18_-_500692 | 0.72 |
ENST00000400256.3 |
COLEC12 |
collectin sub-family member 12 |
| chr1_+_65886326 | 0.72 |
ENST00000371059.3 ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR |
leptin receptor |
| chr4_-_90758118 | 0.70 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr20_+_31350184 | 0.69 |
ENST00000328111.2 ENST00000353855.2 ENST00000348286.2 |
DNMT3B |
DNA (cytosine-5-)-methyltransferase 3 beta |
| chr1_-_85155939 | 0.68 |
ENST00000603677.1 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr14_+_101193164 | 0.68 |
ENST00000341267.4 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chr6_+_44191290 | 0.67 |
ENST00000371755.3 ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr14_+_101193246 | 0.66 |
ENST00000331224.6 |
DLK1 |
delta-like 1 homolog (Drosophila) |
| chrX_-_140271249 | 0.66 |
ENST00000370526.2 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
| chr1_-_47697387 | 0.63 |
ENST00000371884.2 |
TAL1 |
T-cell acute lymphocytic leukemia 1 |
| chr11_+_129939779 | 0.63 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr3_-_88108192 | 0.62 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr3_-_88108212 | 0.61 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr1_-_38471156 | 0.61 |
ENST00000373016.3 |
FHL3 |
four and a half LIM domains 3 |
| chr15_-_83316711 | 0.60 |
ENST00000568128.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr10_-_75255668 | 0.60 |
ENST00000545874.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr11_+_129939811 | 0.59 |
ENST00000345598.5 ENST00000338167.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr13_+_110959598 | 0.58 |
ENST00000360467.5 |
COL4A2 |
collagen, type IV, alpha 2 |
| chr1_+_51434357 | 0.58 |
ENST00000396148.1 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
| chr6_-_110500905 | 0.58 |
ENST00000392587.2 |
WASF1 |
WAS protein family, member 1 |
| chr6_+_134274322 | 0.57 |
ENST00000367871.1 ENST00000237264.4 |
TBPL1 |
TBP-like 1 |
| chr11_-_65640198 | 0.56 |
ENST00000528176.1 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
| chr6_-_112575687 | 0.56 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr11_-_32456891 | 0.55 |
ENST00000452863.3 |
WT1 |
Wilms tumor 1 |
| chr14_-_69445793 | 0.54 |
ENST00000538545.2 ENST00000394419.4 |
ACTN1 |
actinin, alpha 1 |
| chr7_-_150675372 | 0.54 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr9_-_124991124 | 0.53 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr19_-_18548962 | 0.51 |
ENST00000317018.6 ENST00000581800.1 ENST00000583534.1 ENST00000457269.4 ENST00000338128.8 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
| chr19_-_18548921 | 0.51 |
ENST00000545187.1 ENST00000578352.1 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
| chr9_+_96338647 | 0.51 |
ENST00000359246.4 |
PHF2 |
PHD finger protein 2 |
| chr18_+_3449695 | 0.51 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr22_+_38864041 | 0.50 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr6_-_112575838 | 0.50 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr7_-_150652924 | 0.50 |
ENST00000330883.4 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr6_-_110501200 | 0.49 |
ENST00000392586.1 ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1 |
WAS protein family, member 1 |
| chr17_-_8534031 | 0.49 |
ENST00000411957.1 ENST00000396239.1 ENST00000379980.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chrX_+_117480036 | 0.49 |
ENST00000371822.5 ENST00000254029.3 ENST00000371825.3 |
WDR44 |
WD repeat domain 44 |
| chr15_+_39873268 | 0.48 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr17_-_8534067 | 0.48 |
ENST00000360416.3 ENST00000269243.4 |
MYH10 |
myosin, heavy chain 10, non-muscle |
| chr13_-_110959478 | 0.48 |
ENST00000543140.1 ENST00000375820.4 |
COL4A1 |
collagen, type IV, alpha 1 |
| chr20_+_44034676 | 0.48 |
ENST00000372723.3 ENST00000372722.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr5_-_176924562 | 0.47 |
ENST00000359895.2 ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr2_-_158732340 | 0.47 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr1_+_45205498 | 0.46 |
ENST00000372218.4 |
KIF2C |
kinesin family member 2C |
| chr2_-_183903133 | 0.46 |
ENST00000361354.4 |
NCKAP1 |
NCK-associated protein 1 |
| chr1_-_153599732 | 0.46 |
ENST00000392623.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr1_-_153599426 | 0.45 |
ENST00000392622.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr1_+_45205478 | 0.44 |
ENST00000452259.1 ENST00000372224.4 |
KIF2C |
kinesin family member 2C |
| chr19_-_54693521 | 0.44 |
ENST00000391754.1 ENST00000245615.1 ENST00000431666.2 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chr6_-_42016385 | 0.43 |
ENST00000502771.1 ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3 |
cyclin D3 |
| chr7_+_100199800 | 0.43 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chr8_-_27469196 | 0.43 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr6_-_31697977 | 0.43 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_3070313 | 0.43 |
ENST00000326577.4 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chrX_-_55057403 | 0.42 |
ENST00000396198.3 ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2 |
aminolevulinate, delta-, synthase 2 |
| chr7_-_27213893 | 0.42 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr18_+_3449821 | 0.40 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr8_+_55370487 | 0.40 |
ENST00000297316.4 |
SOX17 |
SRY (sex determining region Y)-box 17 |
| chr1_-_202129704 | 0.39 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr16_+_3070356 | 0.39 |
ENST00000341627.5 ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr12_-_96794330 | 0.39 |
ENST00000261211.3 |
CDK17 |
cyclin-dependent kinase 17 |
| chr16_-_30134524 | 0.38 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr20_-_3996165 | 0.38 |
ENST00000545616.2 ENST00000358395.6 |
RNF24 |
ring finger protein 24 |
| chr6_-_112575758 | 0.38 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr11_-_64512803 | 0.37 |
ENST00000377489.1 ENST00000354024.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr18_+_32556892 | 0.37 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_65346555 | 0.37 |
ENST00000542895.1 ENST00000556626.1 |
SPTB |
spectrin, beta, erythrocytic |
| chr9_-_132597529 | 0.36 |
ENST00000372447.3 |
C9orf78 |
chromosome 9 open reading frame 78 |
| chr12_+_6875519 | 0.36 |
ENST00000389462.4 ENST00000540874.1 ENST00000309083.6 |
PTMS |
parathymosin |
| chr1_+_212458834 | 0.36 |
ENST00000261461.2 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_178995021 | 0.35 |
ENST00000263733.4 |
FAM20B |
family with sequence similarity 20, member B |
| chr22_+_19701985 | 0.35 |
ENST00000455784.2 ENST00000406395.1 |
SEPT5 |
septin 5 |
| chr11_-_65325203 | 0.34 |
ENST00000526927.1 ENST00000536982.1 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr6_+_99282570 | 0.34 |
ENST00000328345.5 |
POU3F2 |
POU class 3 homeobox 2 |
| chr22_-_29137771 | 0.34 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr7_-_75368248 | 0.34 |
ENST00000434438.2 ENST00000336926.6 |
HIP1 |
huntingtin interacting protein 1 |
| chr17_+_37824700 | 0.34 |
ENST00000581428.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chrX_+_67913471 | 0.34 |
ENST00000374597.3 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
| chr11_-_34938039 | 0.34 |
ENST00000395787.3 |
APIP |
APAF1 interacting protein |
| chr19_+_3572758 | 0.34 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr19_-_13068012 | 0.33 |
ENST00000316939.1 |
GADD45GIP1 |
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chrX_+_107069063 | 0.33 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr9_+_101569944 | 0.33 |
ENST00000375011.3 |
GALNT12 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr1_-_32403903 | 0.33 |
ENST00000344035.6 ENST00000356536.3 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr3_-_32612263 | 0.33 |
ENST00000432458.2 ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1 |
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr11_-_64546202 | 0.33 |
ENST00000377390.3 ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1 |
splicing factor 1 |
| chr1_+_35734562 | 0.33 |
ENST00000314607.6 ENST00000373297.2 |
ZMYM4 |
zinc finger, MYM-type 4 |
| chr19_+_41107249 | 0.33 |
ENST00000396819.3 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr11_-_47447970 | 0.32 |
ENST00000298852.3 ENST00000530912.1 |
PSMC3 |
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr19_-_42806919 | 0.32 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr6_+_44191507 | 0.32 |
ENST00000371724.1 ENST00000371713.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr9_-_80646374 | 0.32 |
ENST00000286548.4 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
| chr19_-_36523709 | 0.32 |
ENST00000592017.1 ENST00000360535.4 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
| chr12_+_49212514 | 0.32 |
ENST00000301050.2 ENST00000548279.1 ENST00000547230.1 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
| chr8_-_27468842 | 0.32 |
ENST00000523500.1 |
CLU |
clusterin |
| chr19_+_51728316 | 0.32 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
| chr9_+_96338860 | 0.32 |
ENST00000375376.4 |
PHF2 |
PHD finger protein 2 |
| chr19_-_49371711 | 0.32 |
ENST00000355496.5 ENST00000263265.6 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr19_-_42806842 | 0.32 |
ENST00000596265.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr15_+_40733387 | 0.32 |
ENST00000416165.1 |
BAHD1 |
bromo adjacent homology domain containing 1 |
| chr9_-_131709858 | 0.32 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr19_-_41859814 | 0.32 |
ENST00000221930.5 |
TGFB1 |
transforming growth factor, beta 1 |
| chr1_+_246887349 | 0.31 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr21_+_17102311 | 0.31 |
ENST00000285679.6 ENST00000351097.5 ENST00000285681.2 ENST00000400183.2 |
USP25 |
ubiquitin specific peptidase 25 |
| chr8_-_27468945 | 0.31 |
ENST00000405140.3 |
CLU |
clusterin |
| chr10_-_62761188 | 0.31 |
ENST00000357917.4 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr15_-_83316254 | 0.30 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr6_-_32157947 | 0.30 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr9_-_15510287 | 0.30 |
ENST00000397519.2 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr20_+_53092123 | 0.29 |
ENST00000262593.5 |
DOK5 |
docking protein 5 |
| chr5_+_131593364 | 0.29 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr20_-_3996036 | 0.29 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr1_-_161147275 | 0.29 |
ENST00000319769.5 ENST00000367998.1 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr9_+_103235365 | 0.29 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr7_-_108096765 | 0.29 |
ENST00000379024.4 ENST00000351718.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr15_+_91446157 | 0.29 |
ENST00000559717.1 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
| chr12_-_56122426 | 0.28 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr17_+_37824217 | 0.28 |
ENST00000394246.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr6_-_41909191 | 0.28 |
ENST00000512426.1 ENST00000372987.4 |
CCND3 |
cyclin D3 |
| chr3_+_37903432 | 0.28 |
ENST00000443503.2 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_29819096 | 0.28 |
ENST00000568411.1 ENST00000563012.1 ENST00000562557.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr21_+_22370608 | 0.28 |
ENST00000400546.1 |
NCAM2 |
neural cell adhesion molecule 2 |
| chr19_-_3801789 | 0.28 |
ENST00000590849.1 ENST00000395045.2 |
MATK |
megakaryocyte-associated tyrosine kinase |
| chr1_-_16302608 | 0.27 |
ENST00000375743.4 ENST00000375733.2 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
| chr1_+_201924619 | 0.27 |
ENST00000367287.4 |
TIMM17A |
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr5_-_176900610 | 0.27 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chr2_+_66662510 | 0.27 |
ENST00000272369.9 ENST00000407092.2 |
MEIS1 |
Meis homeobox 1 |
| chr1_-_43833628 | 0.27 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr17_+_37824411 | 0.27 |
ENST00000269582.2 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr19_-_10530784 | 0.27 |
ENST00000593124.1 |
CDC37 |
cell division cycle 37 |
| chrX_+_16964794 | 0.26 |
ENST00000357277.3 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr2_-_165477971 | 0.26 |
ENST00000446413.2 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr17_+_57697216 | 0.26 |
ENST00000393043.1 ENST00000269122.3 |
CLTC |
clathrin, heavy chain (Hc) |
| chr10_-_15210615 | 0.26 |
ENST00000378150.1 |
NMT2 |
N-myristoyltransferase 2 |
| chr3_-_47823298 | 0.26 |
ENST00000254480.5 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr6_+_31633902 | 0.25 |
ENST00000375865.2 ENST00000375866.2 |
CSNK2B |
casein kinase 2, beta polypeptide |
| chr7_-_108096822 | 0.25 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr19_+_17666403 | 0.25 |
ENST00000252599.4 |
COLGALT1 |
collagen beta(1-O)galactosyltransferase 1 |
| chr11_-_32457176 | 0.25 |
ENST00000332351.3 |
WT1 |
Wilms tumor 1 |
| chr3_+_101546827 | 0.25 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr6_-_4135825 | 0.25 |
ENST00000380118.3 ENST00000413766.2 ENST00000361538.2 |
ECI2 |
enoyl-CoA delta isomerase 2 |
| chr4_-_57976544 | 0.25 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chr15_-_43785303 | 0.24 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr15_-_89089860 | 0.24 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
| chr1_-_68299130 | 0.24 |
ENST00000370982.3 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
| chr13_-_77460525 | 0.24 |
ENST00000377474.2 ENST00000317765.2 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
| chr11_-_65325430 | 0.24 |
ENST00000322147.4 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chrX_-_53350522 | 0.24 |
ENST00000396435.3 ENST00000375368.5 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
| chr19_-_55919087 | 0.24 |
ENST00000587845.1 ENST00000589978.1 ENST00000264552.9 |
UBE2S |
ubiquitin-conjugating enzyme E2S |
| chr7_-_152133059 | 0.24 |
ENST00000262189.6 ENST00000355193.2 |
KMT2C |
lysine (K)-specific methyltransferase 2C |
| chr3_+_39093481 | 0.24 |
ENST00000302313.5 ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48 |
WD repeat domain 48 |
| chr12_+_49372251 | 0.24 |
ENST00000293549.3 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
| chr5_-_132112921 | 0.23 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chr14_+_29234870 | 0.23 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chr12_-_56120865 | 0.23 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr5_-_115177247 | 0.23 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr5_-_132112907 | 0.23 |
ENST00000458488.2 |
SEPT8 |
septin 8 |
| chr7_-_32931387 | 0.23 |
ENST00000304056.4 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
| chr1_+_27648709 | 0.23 |
ENST00000608611.1 ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222 |
transmembrane protein 222 |
| chr19_-_42806723 | 0.23 |
ENST00000262890.3 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr1_-_182361327 | 0.23 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr18_-_812231 | 0.23 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr5_-_132113083 | 0.23 |
ENST00000296873.7 |
SEPT8 |
septin 8 |
| chr20_+_44034804 | 0.22 |
ENST00000357275.2 ENST00000372720.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr1_+_27648648 | 0.22 |
ENST00000374076.4 |
TMEM222 |
transmembrane protein 222 |
| chr20_+_53092232 | 0.22 |
ENST00000395939.1 |
DOK5 |
docking protein 5 |
| chr2_+_111878483 | 0.22 |
ENST00000308659.8 ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11 |
BCL2-like 11 (apoptosis facilitator) |
| chr8_-_101963482 | 0.22 |
ENST00000419477.2 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr14_+_77228532 | 0.22 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr16_-_28621298 | 0.22 |
ENST00000566189.1 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 1.0 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.3 | 1.4 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.3 | 0.8 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.0 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.2 | 1.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.9 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.2 | 0.6 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.8 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.2 | 1.1 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 1.0 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.2 | 0.3 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.2 | 0.5 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.2 | 2.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 2.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.5 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.2 | 0.5 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.1 | 1.7 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.1 | 0.6 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 1.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.4 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.5 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 1.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.7 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.5 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.6 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 1.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.5 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 1.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.7 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.4 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.4 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.1 | 0.2 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.2 | GO:1900060 | glucosylceramide biosynthetic process(GO:0006679) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.3 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.3 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.1 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.1 | 0.5 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.1 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 2.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.2 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.7 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.3 | GO:0090135 | actin filament branching(GO:0090135) |
| 0.1 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.2 | GO:0032765 | lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 0.2 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.5 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.3 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 1.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 1.8 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.3 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.6 | GO:0051683 | establishment of Golgi localization(GO:0051683) Golgi reassembly(GO:0090168) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.4 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0060592 | mammary gland formation(GO:0060592) |
| 0.0 | 0.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.3 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.0 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 2.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.7 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.3 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 0.9 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.3 | 1.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.0 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.2 | 1.5 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 1.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.5 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.7 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.3 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.2 | GO:0050405 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.1 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.2 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.0 | GO:0070546 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 1.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 0.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.2 | 1.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 1.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.3 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 0.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.2 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 2.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.4 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 2.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 4.0 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 2.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |