Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for EN1_ESX1_GBX1
Z-value: 0.80
Transcription factors associated with EN1_ESX1_GBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
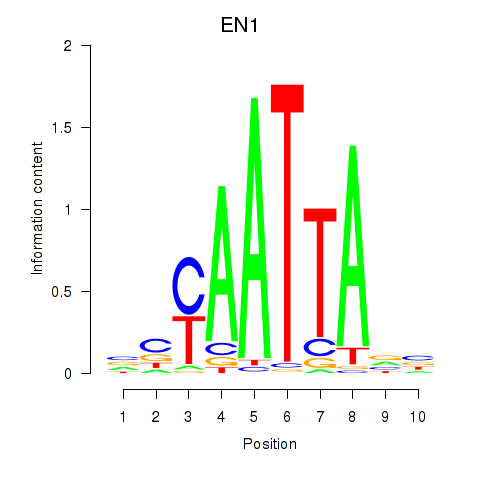
EN1
|
ENSG00000163064.6 | EN1 |
|
ESX1
|
ENSG00000123576.5 | ESX1 |
|
GBX1
|
ENSG00000164900.4 | GBX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EN1 | hg19_v2_chr2_-_119605253_119605264 | 0.28 | 3.0e-01 | Click! |
| GBX1 | hg19_v2_chr7_-_150864635_150864785 | 0.20 | 4.6e-01 | Click! |
Activity profile of EN1_ESX1_GBX1 motif
Sorted Z-values of EN1_ESX1_GBX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EN1_ESX1_GBX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 1.57 |
ENST00000550758.1 |
DCN |
decorin |
| chr13_-_36788718 | 1.24 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr11_-_111794446 | 1.22 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr10_+_5005598 | 1.17 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr12_+_28410128 | 1.15 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr10_-_5046042 | 1.01 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr12_+_26348246 | 0.93 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr7_-_137028534 | 0.93 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr10_-_93392811 | 0.88 |
ENST00000238994.5 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
| chr7_-_137028498 | 0.86 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr8_+_98900132 | 0.84 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr8_+_98881268 | 0.83 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr2_-_145277569 | 0.83 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_-_238323007 | 0.81 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr2_-_238322770 | 0.80 |
ENST00000472056.1 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr2_-_238322800 | 0.79 |
ENST00000392004.3 ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr3_-_123339343 | 0.78 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr5_+_31193847 | 0.75 |
ENST00000514738.1 ENST00000265071.2 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr5_+_135394840 | 0.72 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr13_-_38172863 | 0.70 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr5_+_53751445 | 0.69 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr15_-_37393406 | 0.68 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr5_+_52776449 | 0.68 |
ENST00000396947.3 |
FST |
follistatin |
| chr5_-_42811986 | 0.67 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr6_+_26199737 | 0.67 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr5_+_52776228 | 0.65 |
ENST00000256759.3 |
FST |
follistatin |
| chr11_+_59824060 | 0.64 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr5_-_42812143 | 0.64 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr12_+_26348429 | 0.64 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr11_+_59824127 | 0.62 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr7_+_77428066 | 0.59 |
ENST00000422959.2 ENST00000307305.8 ENST00000424760.1 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr8_-_18744528 | 0.59 |
ENST00000523619.1 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr2_-_188419078 | 0.59 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr7_+_138145076 | 0.58 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr1_+_196788887 | 0.58 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr3_-_123339418 | 0.57 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr4_+_169013666 | 0.54 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr8_-_86290333 | 0.53 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr9_+_125132803 | 0.52 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_+_77428149 | 0.52 |
ENST00000415251.2 ENST00000275575.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr7_-_14029283 | 0.52 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr7_-_99717463 | 0.50 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr3_+_152552685 | 0.49 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chr1_-_109935819 | 0.48 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr16_-_55866997 | 0.48 |
ENST00000360526.3 ENST00000361503.4 |
CES1 |
carboxylesterase 1 |
| chr8_+_9413410 | 0.45 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_-_32157947 | 0.44 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr8_-_41166953 | 0.43 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr1_-_190446759 | 0.42 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr15_-_55563072 | 0.42 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr10_-_49813090 | 0.42 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr3_-_55521323 | 0.42 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr7_-_99716952 | 0.41 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_152214098 | 0.40 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr6_+_39760129 | 0.40 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chrX_+_129473916 | 0.40 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr3_+_69812701 | 0.39 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr12_+_106751436 | 0.39 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr12_+_81110684 | 0.38 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr6_-_26199499 | 0.37 |
ENST00000377831.5 |
HIST1H3D |
histone cluster 1, H3d |
| chr6_-_32145861 | 0.37 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr10_-_99447024 | 0.37 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr1_+_28261533 | 0.36 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr6_-_91296602 | 0.36 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr16_+_50300427 | 0.36 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr16_-_29910853 | 0.36 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr16_+_69345243 | 0.35 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr11_-_117748138 | 0.35 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr13_-_67802549 | 0.35 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr1_+_209878182 | 0.35 |
ENST00000367027.3 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr15_+_58702742 | 0.34 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr19_+_39421556 | 0.34 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr7_-_14029515 | 0.33 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr1_+_43291220 | 0.33 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr6_-_26199471 | 0.33 |
ENST00000341023.1 |
HIST1H2AD |
histone cluster 1, H2ad |
| chr14_+_32798547 | 0.32 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr12_-_51422017 | 0.32 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr6_+_135502501 | 0.31 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_-_91296737 | 0.30 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr7_+_12727250 | 0.30 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr16_-_31085514 | 0.30 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr15_-_55562479 | 0.30 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr6_+_31783291 | 0.29 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr1_+_226013047 | 0.29 |
ENST00000366837.4 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr9_+_80912059 | 0.29 |
ENST00000347159.2 ENST00000376588.3 |
PSAT1 |
phosphoserine aminotransferase 1 |
| chr4_-_103749179 | 0.28 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr9_+_125133315 | 0.28 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_23737534 | 0.27 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr11_-_117747434 | 0.27 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr17_-_40288449 | 0.27 |
ENST00000552162.1 ENST00000550504.1 |
RAB5C |
RAB5C, member RAS oncogene family |
| chr15_-_55562582 | 0.26 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr9_-_95166884 | 0.26 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr11_-_117747607 | 0.26 |
ENST00000540359.1 ENST00000539526.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr3_-_187009646 | 0.26 |
ENST00000296280.6 ENST00000392470.2 ENST00000169293.6 ENST00000439271.1 ENST00000392472.2 ENST00000392475.2 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr2_-_224467093 | 0.26 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr6_-_31830655 | 0.26 |
ENST00000375631.4 |
NEU1 |
sialidase 1 (lysosomal sialidase) |
| chr3_+_157154578 | 0.25 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr17_+_42634844 | 0.25 |
ENST00000315323.3 |
FZD2 |
frizzled family receptor 2 |
| chr9_-_95166841 | 0.25 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr14_-_51027838 | 0.25 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_+_196912902 | 0.25 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr17_-_64225508 | 0.25 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr5_+_174151536 | 0.24 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr13_-_36050819 | 0.23 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chrX_-_6453159 | 0.23 |
ENST00000381089.3 ENST00000398729.1 |
VCX3A |
variable charge, X-linked 3A |
| chr1_-_118472216 | 0.23 |
ENST00000369443.5 |
GDAP2 |
ganglioside induced differentiation associated protein 2 |
| chr6_+_26158343 | 0.23 |
ENST00000377777.4 ENST00000289316.2 |
HIST1H2BD |
histone cluster 1, H2bd |
| chr7_-_14028488 | 0.22 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chrX_+_77166172 | 0.22 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_-_28634874 | 0.22 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr4_-_134070250 | 0.22 |
ENST00000505289.1 ENST00000509715.1 |
RP11-9G1.3 |
RP11-9G1.3 |
| chr9_-_21202204 | 0.21 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr6_+_28317685 | 0.21 |
ENST00000252211.2 ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3 |
zinc finger with KRAB and SCAN domains 3 |
| chr3_-_178984759 | 0.21 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr4_-_103749205 | 0.21 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_3818688 | 0.21 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr11_+_60048053 | 0.21 |
ENST00000337908.4 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr19_-_14640005 | 0.21 |
ENST00000596853.1 ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chrX_-_130423200 | 0.20 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr21_-_35899113 | 0.20 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr17_+_42264556 | 0.20 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr10_+_94451574 | 0.20 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr11_-_32457176 | 0.20 |
ENST00000332351.3 |
WT1 |
Wilms tumor 1 |
| chr3_-_141747950 | 0.20 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_+_77593448 | 0.20 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr1_+_118472343 | 0.20 |
ENST00000369441.3 ENST00000349139.5 |
WDR3 |
WD repeat domain 3 |
| chr19_-_46088068 | 0.20 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr17_-_48785216 | 0.20 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr12_-_10978957 | 0.19 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr7_+_138145145 | 0.19 |
ENST00000415680.2 |
TRIM24 |
tripartite motif containing 24 |
| chr14_+_29236269 | 0.19 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr12_+_104680659 | 0.18 |
ENST00000526691.1 ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1 |
thioredoxin reductase 1 |
| chr4_+_71587669 | 0.18 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr1_-_77685084 | 0.18 |
ENST00000370812.3 ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK |
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr2_-_183387430 | 0.18 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr8_+_22424551 | 0.18 |
ENST00000523348.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr6_-_100912785 | 0.18 |
ENST00000369208.3 |
SIM1 |
single-minded family bHLH transcription factor 1 |
| chr1_+_117544366 | 0.18 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr11_+_60048129 | 0.18 |
ENST00000355131.3 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr4_-_57547454 | 0.18 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr4_-_103749105 | 0.18 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr14_+_32798462 | 0.18 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr8_+_94241867 | 0.17 |
ENST00000598428.1 |
AC016885.1 |
Uncharacterized protein |
| chr17_+_40610862 | 0.17 |
ENST00000393829.2 ENST00000546249.1 ENST00000537728.1 ENST00000264649.6 ENST00000585525.1 ENST00000343619.4 ENST00000544137.1 ENST00000589727.1 ENST00000587824.1 |
ATP6V0A1 |
ATPase, H+ transporting, lysosomal V0 subunit a1 |
| chr3_+_158519654 | 0.17 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr2_-_55237484 | 0.17 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr4_-_103748880 | 0.17 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_138312971 | 0.17 |
ENST00000485115.1 ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70 |
centrosomal protein 70kDa |
| chr6_+_34204642 | 0.17 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr14_-_38064198 | 0.17 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chrX_-_153602991 | 0.16 |
ENST00000369850.3 ENST00000422373.1 |
FLNA |
filamin A, alpha |
| chr6_-_109702885 | 0.16 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr2_+_170440844 | 0.16 |
ENST00000260970.3 ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr19_+_45973120 | 0.16 |
ENST00000592811.1 ENST00000586615.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr11_+_57480046 | 0.16 |
ENST00000378312.4 ENST00000278422.4 |
TMX2 |
thioredoxin-related transmembrane protein 2 |
| chr11_-_3818932 | 0.16 |
ENST00000324932.7 ENST00000359171.4 |
NUP98 |
nucleoporin 98kDa |
| chr4_-_103749313 | 0.16 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_167191814 | 0.16 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chrX_-_106243451 | 0.16 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr15_+_89164520 | 0.16 |
ENST00000332810.3 |
AEN |
apoptosis enhancing nuclease |
| chr10_-_49860525 | 0.15 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr19_-_39421377 | 0.15 |
ENST00000430193.3 ENST00000600042.1 ENST00000221431.6 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
| chr3_+_167453493 | 0.15 |
ENST00000295777.5 ENST00000472747.2 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_-_79315112 | 0.15 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr9_-_116840728 | 0.15 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr15_+_89631647 | 0.15 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr2_-_157189180 | 0.15 |
ENST00000539077.1 ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr14_-_36789783 | 0.15 |
ENST00000605579.1 ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP |
MAP3K12 binding inhibitory protein 1 |
| chr12_-_88974236 | 0.14 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chr15_+_89631381 | 0.14 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chrX_+_108779004 | 0.14 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr19_+_48949030 | 0.14 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr1_+_38478378 | 0.14 |
ENST00000373014.4 |
UTP11L |
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr4_+_147096837 | 0.14 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_-_30228678 | 0.13 |
ENST00000261708.4 |
UTP6 |
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_28969517 | 0.13 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr3_+_115342349 | 0.13 |
ENST00000393780.3 |
GAP43 |
growth associated protein 43 |
| chr8_-_18541603 | 0.13 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr19_+_45458503 | 0.13 |
ENST00000337392.5 ENST00000591304.1 |
CLPTM1 |
cleft lip and palate associated transmembrane protein 1 |
| chr1_-_43855444 | 0.13 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr2_+_68962014 | 0.13 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr1_+_62901968 | 0.13 |
ENST00000452143.1 ENST00000442679.1 ENST00000371146.1 |
USP1 |
ubiquitin specific peptidase 1 |
| chr16_-_11036300 | 0.13 |
ENST00000331808.4 |
DEXI |
Dexi homolog (mouse) |
| chr11_-_64684672 | 0.13 |
ENST00000377264.3 ENST00000421419.2 |
ATG2A |
autophagy related 2A |
| chr1_-_43855479 | 0.13 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chrX_-_20236970 | 0.13 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_-_190927447 | 0.12 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr17_+_79650962 | 0.12 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr2_-_136678123 | 0.12 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr4_-_57547870 | 0.12 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr5_+_125758813 | 0.12 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr2_-_77749474 | 0.12 |
ENST00000409093.1 ENST00000409088.3 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
| chr15_+_62853562 | 0.12 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr18_-_33709268 | 0.12 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr11_-_77790865 | 0.12 |
ENST00000534029.1 ENST00000525085.1 ENST00000527806.1 ENST00000528164.1 ENST00000528251.1 ENST00000530054.1 |
NDUFC2 NDUFC2-KCTD14 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa NDUFC2-KCTD14 readthrough |
| chr1_+_214161272 | 0.11 |
ENST00000498508.2 ENST00000366958.4 |
PROX1 |
prospero homeobox 1 |
| chr6_-_26659913 | 0.11 |
ENST00000480036.1 ENST00000415922.2 |
ZNF322 |
zinc finger protein 322 |
| chr5_-_133968529 | 0.11 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr17_-_40729681 | 0.11 |
ENST00000590760.1 ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP |
PSMC3 interacting protein |
| chr2_+_168725458 | 0.11 |
ENST00000392690.3 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.3 | 0.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 2.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.8 | GO:0060028 | planar cell polarity pathway involved in axis elongation(GO:0003402) convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 1.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.4 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 1.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.3 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.5 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 1.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0034398 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) telomere tethering at nuclear periphery(GO:0034398) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.3 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.1 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.1 | 0.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.2 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.5 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 1.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0036482 | peptidyl-cysteine oxidation(GO:0018171) enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 2.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.8 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 1.0 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:0060926 | atrioventricular node development(GO:0003162) cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 1.3 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.9 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.0 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.7 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 1.0 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.2 | GO:0001657 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 2.2 | GO:0047086 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.3 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.1 | 1.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.3 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 2.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 4.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |